Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 4:01 PM
|
MicrobiomeNet (https://microbiomenet.com) is a web-based platform developed to provide functional insights into microbiome signatures using genome-scale metabolic models (GEMs). It currently hosts 12,400 GEMs and around 6 million microbial signatures. Users can start by searching microbes, metabolites, genes, or enzymes, and perform common tasks such as to characterize the metabolic capacity for a given microbe, to explore known microbial associations, as well as to understand potential metabolic interactions. This book chapter provides practical, step-by-step instructions for navigating MicrobiomeNet to obtain functional insights into individual microbes or microbial association networks. methods

|
Scooped by
mhryu@live.com
Today, 3:49 PM
|
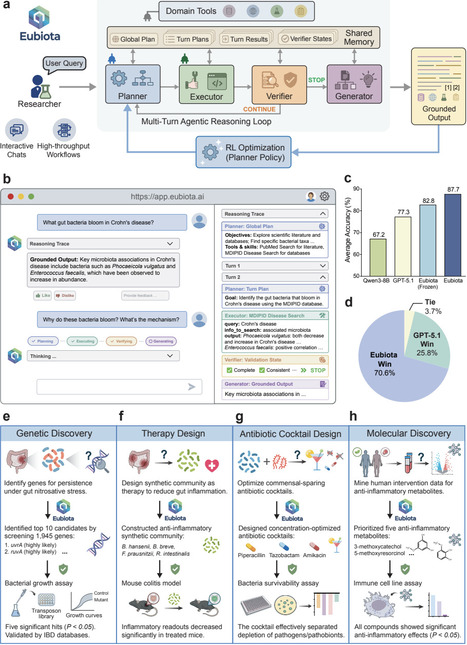
The gut microbiome regulates many aspects of human biology, including immunity and inflammatory diseases, yet mechanistic discovery and translation remain constrained by fragmented workflows and manual hypothesis integration. Here, we present Eubiota, an open-source modular agentic framework that decomposes microbiome inquiry using specialized agents for planning, execution, verification, and grounded synthesis. Coordinated through shared memory and domain-specific tools, Eubiota employs reinforcement learning to optimize multi-turn reasoning, achieving 87.7% benchmark accuracy and outperforming GPT-5.1 by 10.4%. Across four case studies, Eubiota enabled end-to-end discovery with experimental validation. Screening nearly 2,000 bacterial genes, Eubiota identified the uvr-ruv DNA repair axis as a fitness determinant under inflammatory stress, validated using transposon mutants and IBD metagenomes. It further designed a four-strain consortium that attenuated colitis severity in mice and generated a commensal-sparing antibiotic cocktail, demonstrating its utility in addressing community-level design challenges at cellular and molecular levels. Finally, Eubiota discovered diet-associated metabolites that suppress NF-kB signaling. Together, these results establish Eubiota as a scalable, tool-grounded scientific copilot for mechanistically driven microbiome discovery.

|
Scooped by
mhryu@live.com
Today, 10:41 AM
|
The oxygen reduction reaction (ORR) in living cells efficiently generates energy but is limited for sustainable applications due to inefficient electron transfer across membranes. Here, we report an interspecies cooperative mechanism, termed electro-mutualism, that enables efficient extracellular ORR using an electrotrophic Acinetobacter venetianus RAG-1 (RAG-1) and a non-CO2-fixing electrotrophic Shewanella oneidensis MR-1 (MR-1), sustained by inorganic carbon supplied via a bicarbonate–CO2 equilibrium. Under cathodic polarization, RAG-1 assimilates inorganic carbon and secretes lactate using electrode-derived electrons, which fuels MR-1. In turn, MR-1 releases flavins that interact with the RnfB complex of RAG-1, thereby accelerating the transmembrane electron transfer. This electro-mutualistic cooperation achieves, to our knowledge, the highest reported whole-cell ORR current density (20.9 A/m2) under O2-aerated, neutral-pH conditions, outperforming benchmark Pt/C and laccase cathodes evaluated under identical conditions. The universality of electro-mutualism is further supported by replacing MR-1 with Bacillus subtilis and is predicted to extend across proteobacteria, firmicutes, and actinobacteria. Electro-mutualism thus provides a metal-free, genetically unmodified catalytic strategy for green energy conversion.

|
Scooped by
mhryu@live.com
Today, 10:29 AM
|
In recent years, with the rapid advancement of synthetic biology and nanoscience, microbial therapy based on engineered bacteria has garnered renewed widespread attention and achieved remarkable research progress. Specifically, through biological and physicochemical methods, bacteria from diverse sources with unique characteristics are functionally modified and engineered to combine with conventional therapeutic approaches. This aims to overcome or mitigate the limitations of standard treatments in clinical applications. On this basis, the integration of exogenously responsive regulatory factors (e.g., ultrasound) with microbial therapy undoubtedly provides a more intelligent, precise, and promising therapeutic strategy for tumor treatment. In this review, we summarize the core methods for direct modification, mechanisms of action, and main application strategies of engineered bacteria in tumor therapy. Meanwhile, we focus on elaborating the forms and interaction mechanisms of combining ultrasound with engineered bacteria, and discuss in detail the latest research advances in their combined application for tumor therapy. A comprehensive understanding of their relationships and interaction mechanisms will provide theoretical support for the development of ultrasound-regulated bioactive molecules and ultrasound-responsive intelligent drug delivery systems, thereby further advancing the applications of ultrasound technology and microbial therapy in the biomedical field.

|
Scooped by
mhryu@live.com
February 28, 4:49 PM
|
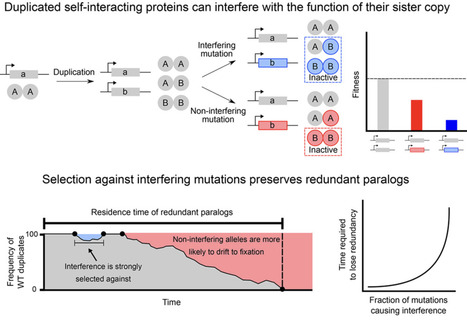
Models of gene duplication often assume that loss-of-function mutations neutrally promote the return to the ancestral singleton state. They thus ignore the potential functional interference between duplicated proteins stemming from their physical interactions. Here, we show that for heteromerizing paralogs, such interference potentiates negative selection on loss-of-function mutations. This effect maintains genetic redundancy over longer timescales depending on the rate and severity of loss-of-function mutations. We experimentally estimate that around 6% of deleterious amino acid substitutions in a representative tetrameric protein interfere with a second copy. Interfering substitutions typically disrupt either catalysis or the final step of protein complex assembly, with varying degrees of severity. Our work shows that paralog interference renders the negative effects of loss-of-function mutations visible to purifying selection, contributing to the preservation of genetic redundancy.

|
Scooped by
mhryu@live.com
February 28, 4:05 PM
|
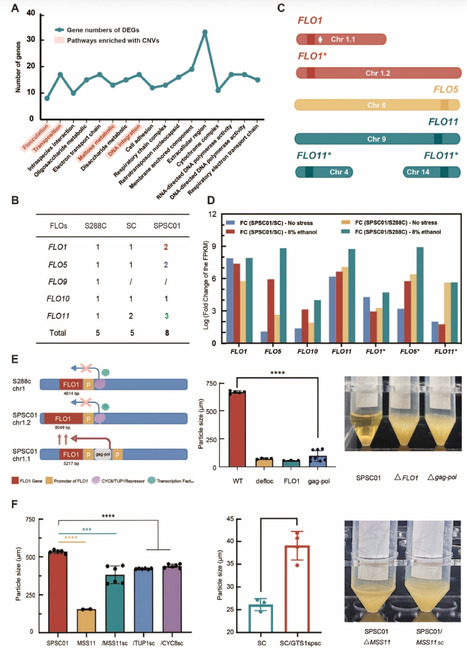
A unique self-flocculating yeast strain SPSC01 was developed through protoplast fusion for fuel ethanol production with high product titers. In this study, we conducted comparative multi-omics analyses on SPSC01 to elucidate mechanisms underlying its self-flocculating phenotype and associated stress tolerance, the most desirable merit for robust production in industry. Leveraging two cutting-edge third-generation sequencing technologies, we achieved a gapless high-quality and chromosome-level assembly for the genomes of SPSC01 and its parental strains. Through comprehensive genome analyses, we identified 25 unique genes that are absent in the parental strains, along with 13 novel genes with unknown functions. The self-flocculation of yeast cells is driven by the copy number of genetic variations and significantly upregulated transcription of FLO genes. Mutations in both cis- and trans-regulatory elements contribute to the constitutive expression of FLO1 and its derivative genes, a prerequisite for developing the self-flocculating phenotype. Notably, we discovered a novel small protein G12 that harbors a zinc finger domain, and its overexpression substantially enhanced ethanol production of engineered yeast strains. Furthermore, alterations in metabolic pathways with ergosterol, glutathione, amino acid, and glycerophospholipid are implicated for developing tolerance to ethanol and major inhibitors acetic acid and furfural that are released during the pretreatment of lignocellulosic biomass. The progress provides strategies for engineering yeast cell factories with robustness through rational design to produce biofuels and bio-based chemicals with high product titers and productivities, in particular with the biorefinery of lignocellulosic biomass for sustainable socioeconomic development.

|
Scooped by
mhryu@live.com
February 28, 3:29 PM
|
Terpenes constitute a vast and industrially important class of natural products. Yet, microbial production of many high-value terpenoids remains limited by the difficulty of rationally engineering their biosynthetic pathways and the lack of high-throughput screening systems that directly report product formation. This challenge is especially acute for monoterpene alcohols such as α-terpineol (1), whose biosynthesis in heterologous hosts requires coordinated precursor formation, cyclization, and water-capture chemistry. Here, we develop a transcription factor–based whole-cell biosensor strain capable of detecting 1 by engineering the p-cumate repressor CymR through structure-guided directed evolution. Guided by a model of the putative ligand-binding pocket, focused libraries at residues implicated in effector accommodation yielded variants with dramatically improved sensitivity. This culminated in the CymR variant 3-A8, which exhibits a 22-fold increase in dynamic range relative to wild-type. Using this optimized biosensor, we demonstrate in vivo monitoring of 1 production in E. coli by coupling it to an artificial alcohol-dependent hemiterpene (ADH) pathway and downstream modules expressing GPPS and α-terpineol synthase. The integrated biosensor–production system effectively distinguishes the complete biosynthetic pathway from deletions and reports intracellular titers consistent with GC-MS quantification. Together, these results provide the first biosensor for monocyclic monoterpene alcohols and establish a compact, modular framework for high-throughput screening and pathway optimization. This platform sets the stage for accelerating the discovery, engineering, and scalable bioproduction of valuable isoprenoids and other terpene-derived natural products.

|
Scooped by
mhryu@live.com
February 28, 3:11 PM
|
Recent advances in Artificial Intelligence (AI) are reshaping kinase research by uncovering complex regulatory mechanisms and accelerating drug discovery. These advances have enabled the capture of elusive allosteric transitions and transient cryptic pockets by exploring dynamic conformational landscapes, revealing features critical for understanding ligand interactions and enzymatic regulation. AI-driven structure prediction tools, such as AlphaFold (AF)-based models, offer high-resolution insights into kinase conformations and complex assembly, thereby clarifying receptor activation in signaling pathways. Moreover, cutting-edge AI frameworks provide fresh perspectives on substrate prediction, cofactor mapping, and mutation-induced structural changes. Integrative strategies that combine Machine Learning (ML), molecular docking, Molecular Dynamics (MD) simulation, and experimental validation have further streamlined the discovery of novel kinase inhibitors and related studies. Collectively, these AI-enhanced approaches deepen our understanding of kinase-mediated signaling—from developmental processes to oncogenic transformation—and highlight the powerful synergy between computational and experimental research in advancing therapeutic innovations.

|
Scooped by
mhryu@live.com
February 28, 2:52 PM
|
The Type VI secretion system (T6SS) is widely recognized as a contractile nanomachine that mediates interbacterial antagonism, yet its biological roles and evolutionary logic vary substantially across bacterial lineages. In this Review, we synthesize recent advances in the Acinetobacter T6SS field and propose a unifying perspective in which the system functions as a context-dependent fitness module rather than a constitutive virulence weapon. We highlight how Acinetobacter has rewired a single T6SS platform through non-canonical structural solutions, multilayered regulatory integration, and an unusually expansive effector repertoire. Beyond microbial competition, emerging clinical and experimental evidence links T6SS activity to host immune amplification, disease severity, and the dynamics of horizontal gene transfer and antibiotic resistance. By integrating structural biology, regulatory logic, effector function, and clinical observations, this Review reframes the Acinetobacter T6SS as an adaptable system that balances aggression, persistence, and metabolic cost in polymicrobial and host-associated environments. This perspective not only advances conceptual understanding of T6SS diversity but also highlights translational opportunities for diagnostics, vaccines, and anti-virulence strategies targeting multidrug-resistant Acinetobacter infections. We synthesize advances in Acinetobacter T6SS biology, revealing how non-canonical structural solutions and multilayered regulation create an adaptable system for competition, persistence, and host immune modulation.

|
Scooped by
mhryu@live.com
February 28, 2:33 PM
|
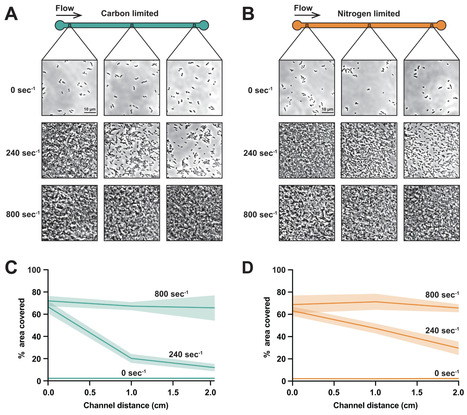
In laboratory settings, bacteria grow in static culture with more nutrients than they require. However, bacteria in nature experience flowing environments that are nutrient-limited. Using microfluidics and single-cell imaging, we discover that shear flow promotes growth of the human pathogens Pseudomonas aeruginosa and Vibrio cholerae at surprisingly low nutrient concentrations. In static environments, cells require high nutrient concentrations as they steadily consume non-renewable resources. In slower-flowing environments, cells grow and deplete nutrients, which generates spatial gradient profiles. In faster-flowing environments, cells grow robustly and form microcolonies even at very low concentrations due to rapid nutrient replenishment. By precisely delivering nutrients using microfluidics, we learned that cells in flow can grow on glucose concentrations 1,000 times lower than those observed in typical laboratory experiments. The ultralow glucose concentrations sufficient for growth in flow closely align with the affinity of bacterial glucose transporters, suggesting that bacteria have evolved in flowing environments with scarce nutrients. Collectively, our results emphasize the limits of traditional culturing approaches and highlight how shear flow can promote bacterial growth and shape spatial gradients.

|
Scooped by
mhryu@live.com
February 28, 2:13 PM
|
The gut microbiome and its effects on human health have generated considerable scientific, veterinary, and medical interest in recent years. Several gut bacterial species have emerged as potential chassis organisms for the delivery of therapeutics in this milieu. Among these, E. coli Nissle 1917 (EcN), a nonpathogenic gut isolate bacterium, is quickly gaining popularity. However, a bottleneck in harnessing EcN’s potential has been its poor transformation efficiency relative to other bacterial strains. In this study, we present the use of adaptive laboratory evolution to increase EcN’s transformation efficiency by subjecting the strain to repeated cycles of electroporation and recovery. This new strain has been comprehensively characterized in comparison to the wild-type EcN, including assessments of growth under gut-mimicking duress conditions, permeability, motility, hydrophobicity, and plasmid replication. Since EcN is known to compete with pathogenic strains in the gut for iron, the competition dynamics and iron consumption of the strain were also significant factors to consider. Furthermore, we conducted genome sequencing and gene ontology enrichment analysis to identify affected genes and pathways to probe the potential mechanisms of the improved phenotype. Overall, the strain shows improved transformation efficiency and robustness while preserving its key biological functionality.

|
Scooped by
mhryu@live.com
February 28, 2:05 PM
|
Multiplex genome editing (MGE) enables coordinated modification of multiple genomic loci and is foundational for engineering complex biological traits. Traditional CRISPR-Cas nuclease-based strategies rely on DNA double-strand breaks (DSBs), which limit precision and pose scaling challenges for incorporating simultaneous edits across different loci. Recent advances in genome editing technologies that operate without generating DSBs have expanded the accuracy and feasibility of multiplexed genomic manipulation. This review focuses on emerging strategies for precise MGE, including base editing, prime editing, and related genome rewriting platforms. We highlight key engineering principles that impact the success of scalable multiplexing, including the choice of editing platform, edit size, and guide RNA architecture, and discuss applications across mammalian, plant, fungal, and bacterial systems. Together, these technologies establish MGE as a versatile framework for precise multigene control in biotechnology and agriculture.

|
Scooped by
mhryu@live.com
February 28, 1:37 PM
|
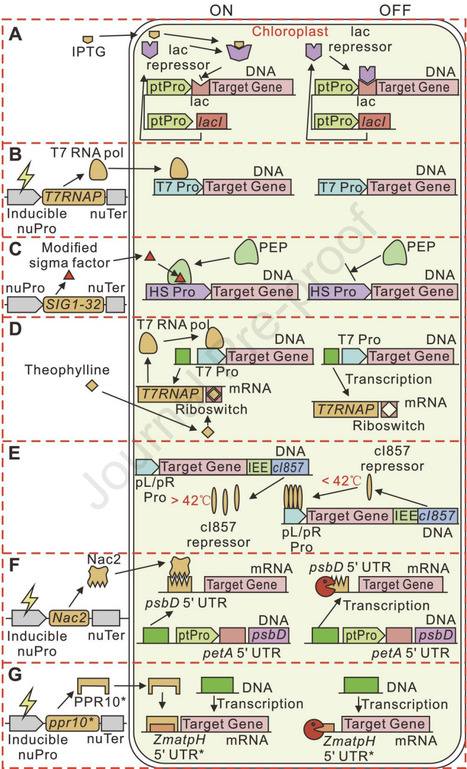
Chloroplasts are central to photosynthesis and metabolic biosynthesis in plants and algae, offering immense potential for synthetic biology. This review highlights recent advances in chloroplast genome minimization, high-efficiency genome editing, and metabolic pathway redesign to optimize chloroplast functionality. We begin by summarizing strategies to relocate chloroplast-encoded genes to the nuclear genome, facilitating chloroplast genome streamlining while maintaining gene expression. We then discuss cutting-edge tools for precise chloroplast genome editing, enabling knockout, knock-in, base substitution, and multiplex modifications. Finally, we explore the dynamic regulation of chloroplast energy metabolism and the redesign of core pathways to establish de novo biosynthesis of high-value compounds, enhancing production efficiency and enabling crop trait improvement. These integrated approaches pave the way for next-generation chloroplast engineering with broad implications for biotechnology and agriculture.
|

|
Scooped by
mhryu@live.com
Today, 3:55 PM
|
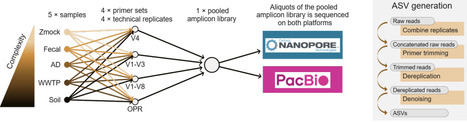
Sequencing of ribosomal marker genes remains a cornerstone for profiling complex microbial communities. In recent years, there has been a shift from Illumina to long-read technologies, including PacBio and Oxford Nanopore Technologies (ONT). ONT is attractive due to its low startup cost and portability; however, historically high error rates have prevented direct amplicon sequencing variant (ASV) generation from raw nanopore reads. This has forced most workflows to rely on mapping raw reads against reference databases constraining analyses to taxa covered by these. With recent improvements in ONT sequencing accuracy, we sought to challenge this view by sequencing samples of increasing complexity using primer sets targeting amplicons of different lengths, and by sequencing the exact same PCR libraries on both PacBio and ONT. We demonstrate that error-free ASVs can now be generated directly from raw nanopore reads using standard denoising algorithms originally developed for Illumina data. Current ONT read quality enables reliable reconstruction of amplicons spanning ~250 bp to ~4,200 bp and allows resolution of intragenomic rRNA gene variants. These results extend beyond simple mock communities to complex fecal, anaerobic digester, activated sludge, and soil samples. When sequencing depth is sufficient, ONT accurately recovers all or nearly all intra-genomic 16S rRNA gene copy variants, showing perfect sequence identity to curated reference sequences in mock communities and to ASVs inferred from PacBio data in complex communities. Across the primer sets, ONT required higher sequencing depth than PacBio to fully resolve the communities, with this requirement increasing with amplicon length. For complex samples, ONT required approximately 2-3x more reads for V4 (~250 bp) and V1-V3 (~500 bp), 4.1-5.6x more reads for V1-V8 (~1400 bp), and 25-42x more reads for rRNA operon (OPR) amplicons (~4200 bp). Consequently, sequencing complex communities with OPR primers on ONT is currently not feasible due to the unrealistically high read depth required. This study provides evidence that ONT amplicon sequencing has matured to the point where true ASV-resolved profiling is practically and economically feasible, moving ONT amplicon analysis beyond reliance on OTU clustering or reference alignment to enable application in environments lacking comprehensive reference databases.

|
Scooped by
mhryu@live.com
Today, 3:36 PM
|
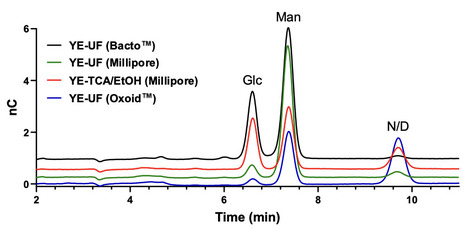
Exopolysaccharides (EPS) produced by lactic acid bacteria (LAB) and other microorganisms have attracted considerable interest due to their structural diversity and physicochemical properties, which makes them valuable across various industrial applications. To achieve high cell densities and maximize EPS yields, microorganisms are typically cultivated in nutrient-rich media containing yeast extract. However, yeast extract may contain high molecular weight polysaccharides that are not metabolized by the bacteria. This can lead to an overestimation of EPS yields and contamination of the bacterial EPS, potentially resulting in misinterpretation of their structure and biological activity. In this study, we demonstrate the presence of high molecular weight α-mannan and β-glucan in yeast extract in EPS isolates using both ultrafiltration and the commonly used trichloroacetic acid/ethanol (TCA/EtOH) precipitation method. These polysaccharides were characterized by size-exclusion chromatography, high-performance anion-exchange chromatography, and nuclear magnetic resonance spectroscopy. Their abundances were estimated to range from 10 to 50 mg/L in MRS medium, depending on the supplier of the yeast extract. The main contaminant identified was yeast α-mannan. By cultivating L. rhamnosus GG (ATCC 53103) and L. pentosus KW1 and isolating their respective EPS, we illustrate how these yeast extract contaminants affect the structural interpretation of the EPS and that the contaminants can be completely removed by ultrafiltration of the growth medium prior to bacterial cultivation. In conclusion, we emphasize the necessity of stringent controls in the production and purification of microbial EPS, with particular attention to the chemical purity of medium constituents.

|
Scooped by
mhryu@live.com
Today, 10:36 AM
|
The persistent global challenge of foodborne pathogens necessitates the development of ultrasensitive point-of-care (POC) detection technologies. However, conventional methods for detecting foodborne pathogens, such as culture-based and nucleic-acid-based techniques, are often limited by complex procedures and reliance on sophisticated instrumentation, respectively. Although immunoassays are more suitable for POC detection, they are frequently constrained by the high cost and limited stability of the antibodies. Therefore, there is an urgent need to develop innovative, ultrasensitive POC biosensing platforms for foodborne pathogens detection. This comprehensive review discusses the development of phage-based nano-sensors for advancing ultrasensitive POC detection of foodborne pathogens. Initially, the inherent properties of phages and phage engineering strategies for creating ideal recognition elements are briefly introduced. Subsequently, phage-based nanosensors for foodborne pathogen detection and their signal amplification mechanisms are comprehensively reviewed, including colorimetric, fluorescent, electrochemical, magnetic relaxation switch, chemiluminescent, and other modalities. Then, the integration of artificial intelligence into intelligent analysis is highlighted. Furthermore, current challenges and future prospects are outlined to bridge the gap between laboratory research and practical application. The integration of phages into nanosensors offers opportunities to overcome the limitations of conventional methods for monitoring foodborne pathogens and is expected to significantly enhance food safety in control systems.

|
Scooped by
mhryu@live.com
Today, 9:52 AM
|
Data storage using proteins offers high capacity and stability, enabling utilization of protein techniques for data storage and retrieval. However, expressing unnatural proteins with random sequences for data storage and sequencing them for accurate data retrieval remain challenging. In this study, by encoding digital data into amino acid sequences and incorporating them into collagen-like protein templates, we achieve successful expression of the proteins via E. coli for data storage; the data-bearing proteins containing selective amino acids and arginine intervals can be sequenced through tryptic digestion followed by LC-MS/MS analysis to achieve complete data recovery, even for protein mixtures encoding multiple datasets. We further demonstrate much higher stability of the data-bearing protein than DNA, and random access and cryptographic data protection using affinity-tagged proteins. This work establishes a robust framework for protein-based data storage, opening up avenues for data storage and retrieval, protein engineering and chemistry, synthetic biology, proteomics, and beyond. Data storage using proteins offers high capacity and stability, however, expressing unnatural proteins with random sequences often fails. Here the authors encode digital data into amino acid sequences based on collagen-like protein templates to allow stable data storage and retrieval.

|
Scooped by
mhryu@live.com
February 28, 4:09 PM
|
The allocation of finite cellular resources is a fundamental principle that dictates microbial metabolic strategies and gives rise to complex phenomena, such as overflow metabolism, characterized by the production of respiro-fermentative by-products, including acetate, during rapid growth. Although proteome-constrained models have successfully predicted overflow metabolism in E. coli, they often overlook the distinct biophysical and energetic costs associated with protein localization. The cellular membrane, in particular, represents a critical and constrained compartment where competition for space and synthesis machinery can create significant metabolic bottlenecks. To investigate this, we developed the membrane-associated constrained flux balance analysis (MAFBA), a scalable, genome-scale metabolic model that introduces a tunable constraint on the total protein mass allocated to the cellular membrane. Our model demonstrates that the overall and membrane-associated proteome allocation constraints interact to improve the accuracy of predicting the onset of overflow metabolism. It mechanistically reveals that at high growth rates, competition for limited membrane allocation forces a trade-off between growth-essential functions and respiratory capacity, leading to acetate production. Furthermore, MAFBA quantitatively explains the widely observed experimental phenomenon that expressing heterologous membrane proteins imposes a significantly higher metabolic burden than expressing cytosolic proteins. This study establishes membrane resource allocation as a key constraint governing bacterial physiology, acting in concert with overall proteome limitations. The resulting MAFBA framework provides a powerful and accessible tool for synthetic biology and metabolic engineering, enabling the prediction of metabolic costs associated with expressing membrane-bound proteins and guiding strain design strategies, holding promise for applications in bioproduction and metabolic engineering.

|
Scooped by
mhryu@live.com
February 28, 3:51 PM
|
Even though complex microbial communities are ubiquitous and provide essential services for natural and human-associated ecosystems, our knowledge about their assembly and dynamics is incomplete. There is an ongoing debate whether the behavior of complex communities can be predicted from the outcome of pairwise competition of species, and whether communities reach alternative stable states depending on the level and complexity of resource provided for growth. To estimate the effect of two resource gradients, total carbon availability and resource complexity, on the compositional dynamics of a complex microbial community, we conducted a 16-day serial passage experiment, transferring a 16-species synthetic community in 96 different resource environments. We observed that although both resource dimensions influenced community composition, total carbon exerted a considerably larger effect. Additionally, we saw the emergence of a tristable pattern along the total carbon gradient, a feature not observed for the resource complexity gradient. Using monoculture assays, we identified lag phase duration as the dominant predictor of competitive success at carbon extremes, with maximum growth rate increasing in importance as lag times converged. Total carbon availability thus structured community state transitions and regulated which growth trait governed competitive sorting. These results suggest the importance of total carbon level over resource complexity and identifying dominant species for the quest to successfully manage, maintain and manipulate complex microbial communities.

|
Scooped by
mhryu@live.com
February 28, 3:17 PM
|
Plant microbiome disruption often increases vulnerability to crop diseases, endangering worldwide food production, while chemical pesticides become increasingly less viable and continue to damage ecosystems. To safeguard plant microbiome health, several biological control strategies offer alternatives, yet many operate through broader or weakly defined target mechanisms. In recent years, bacterial contractile injection systems (BCISs) have emerged as a promising class of naturally evolved nanomachines that translocate molecular payloads directly into target cells. Subsets of these systems, extracellular contractile injection systems (eCISs), are distinguished by their specific narrow host range and receptor-dependent specificity. Recent studies have demonstrated that eCISs provide a transformative approach for targeted microbial manipulation, enabling the delivery of specialized molecules into particular microbes with higher precision. However, despite their potential, the integration of these engineered injection systems with microbial modulation for phytomicrobiome remains largely underexplored. Here, we explore the capabilities of eCISs as an advanced approach for the biocontrol, leveraging their tailored mechanisms for targeted payload delivery in plant-associated microbial communities with enhanced host specificity. This study aims to address the potential of engineered injection systems in facilitating sustainable phytomicrobiome engineering strategies that enhance biocontrol, aiming to reduce environmental harm while improving agricultural productivity.

|
Scooped by
mhryu@live.com
February 28, 2:57 PM
|
Aptamers are conventionally selected via ‘Systematic Evolution of Ligands by Exponential Enrichment’ (SELEX). However, this process is laborious, time-consuming, and has a relatively low efficacy. In this study, we present a novel automated high-throughput screening platform that augments the conventional selection of DNA aptamers. To this end, the software of an optical next-generation sequencer has been modified to automatically perform fluorescence-based binding assays on the displayed DNA sequences subsequent to sequencing. Utilizing this platform, high-affinity DNA aptamers were identified for the proteins LecA, LecB, and Pseudomonas Exotoxin A (PEA) of Pseudomonas aeruginosa following pre-enrichment by a mere three to five SELEX rounds. Conversely, 12 rounds of conventional SELEX yielded aptamers exhibiting three-fold lower affinity for LecA and PEA, with no aptamers obtained for LecB. Furthermore, we demonstrate that the proposed method is suitable for the study of molecules ranging from small molecules to whole cells. This is evidenced by a mutation assay for a kanamycin-binding aptamer and the monitoring of E. coli binding to aptamers. The present study proposes a high-throughput approach to enhance SELEX, with the potential to provide greater insight into the selection process and to significantly increase efficacy, enabling the selection of aptamers within a week.

|
Scooped by
mhryu@live.com
February 28, 2:48 PM
|
Large amounts of waste salt formation are a major bottleneck in the industrial production of l-lactic acid (l-LA). In this study, we developed a cost-effective, neutralizer-free process with industrial potential using acid-tolerant yeast Pichia kudriavzevii. Based on the previously engineered P. kudriavzevii Pkz04, we further improved l-LA production by inserting multicopy l-lactate dehydrogenase, performing promoter replacement, and introducing a lactate transporter, resulting in 102.32 g/l l-LA without neutralizers. Subsequently, we performed batch fermentation in a low-cost inorganic salt medium containing only 0.03 g/l yeast extract and achieved 75.09 g/l l-LA, which outperformed most reported strains with respect to a yield of 0.99 g/g glucose. Finally, a detailed technoeconomic analysis was conducted, including an analysis of low concentrations of organic nitrogen sources, pH-adjustment and nonadjustment processes, and carbon losses in different fermentation processes. Our results highlight the potential of low-pH production of l-LA from an industrial perspective, which is both financially viable and environmentally beneficial.

|
Scooped by
mhryu@live.com
February 28, 2:29 PM
|
RNA isolation is a critical first step for gene expression analysis; however, obtaining high-quality RNA from polysaccharide-rich or biofilm-producing microbial samples remains challenging. High polysaccharide content hinders cell lysis, decreases RNA yield, and reduces sample purity, limiting the reliability and accuracy of downstream techniques such as RNA-seq and RT-qPCR. In this study, we evaluated the use of polysaccharide lyase, Smlt1473, as a pre-processing step to improve RNA isolation from Pseudomonas species. Incorporation of Smlt1473 into a commercial RNA extraction kit workflow significantly improved RNA extraction for mucoid clinical and agricultural pseudomonads and did not adversely affect the non-mucoid isolate, PA14. More specifically, RNA-seq analysis demonstrated that pre-processing with Smlt1473 increased the proportion of assigned reads without introducing significant changes in gene expression. Only a limited set of genes—primarily hypothetical proteins and potential phage-associated elements—were differentially expressed, while global transcriptional profiles remained stable. Together, these findings demonstrate that pre-processing with Smlt1473 provides an effective, easily integrated method to enhance RNA isolation from biofilm-forming bacteria and improves RNA-seq data quality without perturbing the underlying biology.

|
Scooped by
mhryu@live.com
February 28, 2:10 PM
|
Collectively known as one-carbon metabolism (OCM), both the folate and methionine cycles are highly regulated to meet cellular demands. These cycles are key in the production and recycling of methyl groups to be used in many essential cellular processes such as the production of nucleotides, as well as S-adenosyl-l-methionine (SAM) the global methyl donor for DNA, RNA, and post translational modifications. Within the folate cycle, 5,10-methylenetetrahydrofolate is the main species through which methyl groups enter OCM. Therefore, 5,10-methylenetetrahydrofolate reductase (MTHFR), which reduces 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, is the central enzyme that directs methyl groups for use within the methionine cycle. MTHFR is an enzyme found in all domains of life, but unlike in prokaryotes, eukaryotic MTHFR activity is highly regulated by the level of SAM, to balance the one-carbon needs of the cell. In this perspective, we review the catalytic mechanism of MTHFR, evolutionary differences, and the regulatory mechanisms that have evolved to alter its activity. We also discuss recent structural findings that reveal a unique mechanism for inactivation by SAM as a feedback loop and its consequences for understanding inherited MTHFR deficiency.

|
Scooped by
mhryu@live.com
February 28, 2:03 PM
|
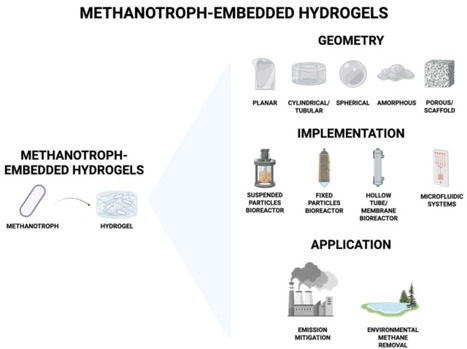
Methane is a potent greenhouse gas with a short atmospheric lifetime, making its removal critical for mitigating climate change. Methanotrophic microorganisms enzymatically oxidize methane at ambient temperatures, offering a biological route for mitigation. However, conventional liquid-based bioreactors face mass transfer limitations and scalability challenges. Here, we review studies of methanotrophic biofilms and explore their embedding within hydrogel matrices, which can enhance gas diffusion, support stable biofilm growth, and improve methane uptake rates, as a biotechnology for methane removal. These hydrogel-embedded methanotrophic systems combine biocompatibility and structural advantages to facilitate efficient methane uptake and removal. This approach holds promise for scalable, cost-effective bioreactors capable of reducing methane emissions from diverse sources, thereby contributing to near-term climate change mitigation efforts.
|
 Your new post is loading...
Your new post is loading...