 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 4:46 PM
|
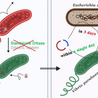
The pathogenic bacterium Vibrio parahaemolyticus represents a substantial economic and public health concern; however, elucidating its virulence mechanisms has been significantly impeded by its inherent resistant to genetic manipulation, primarily attributed to sophisticated immune defense systems including restriction-modification (R-M) modules, CRISPR-Cas systems, standalone DNases, and DdmDE systems. Paradoxically, while genetic modification is essential for overcoming these barriers, the very barriers themselves obstruct DNA introduction. Our investigation focused on the V. parahaemolyticus X1 strain, where initial plasmid transformation attempts proved unsuccessful. However, low-efficiency conjugation allowed knockout of defense genes, thereby silencing the host’s defense mechanisms. Our findings revealed a standalone DNase, Vpn, as the predominant obstacle to foreign DNA entry in the X1 strain, while a DdmDE system executes elimination of invaded plasmids. Leveraging these insights, we created the V. parahaemolyticus X2 strain via sequential depletion of the Vpn nuclease and the DdmDE system. Capitalizing on the bacterium’s exceptional growth rate, characterized by a generation time of approximately 10.5 min, we established a highly efficient molecular cloning platform capable of creating a new plasmid construct within a single day. This work not only presents a strategic framework for genetic manipulation of previously recalcitrant bacterial species but also underscores the potential of fast-growing marine bacteria as promising candidates for next-generation biotechnological applications.

|
Scooped by
mhryu@live.com
Today, 4:39 PM
|
Single-cell resolution studies have transformed our understanding of microbial systems, revealing substantial cell-to-cell heterogeneity and complex dynamic behaviors. This review describes recent advances in using optogenetics, where light-sensitive proteins control cellular processes, to investigate microbial behavior at the individual cell level. We discuss studies where optogenetic approaches have enabled high-resolution analysis of properties such as relative cell positioning, subcellular localization, morphology, and gene expression dynamics. In addition, we highlight emerging feedback and event-driven control methods that dynamically modulate cellular states using light signals. By leveraging light's unique capabilities for spatial and temporal manipulation, researchers can now probe cellular characteristics with unprecedented precision. We anticipate significant advances as researchers introduce more sophisticated dynamically patterned light signals for single-cell microbial research.

|
Scooped by
mhryu@live.com
Today, 4:34 PM
|
Magnetotactic bacteria (MTB) utilize magnetosomes to align passively with Earth's magnetic field. Magnetic alignment, coupled with flagellar motility and aerotaxis, enables MTB to perform magneto-aerotaxis-a strategy that constrains their movement to a one-dimensional trajectory along geomagnetic field lines, optimizing their search for low-oxygen niches in aquatic environments. Beyond axially constrained movement, environmental MTB isolates exhibit a hemispherically determined swimming polarity-favoring either magnetic north or south-that has been suggested to facilitate descent into oxygen-depleted zones. However, a direct quantitative evaluation of how matching swimming polarity influences navigation toward low-oxygen environments has remained elusive. Here, we employed microcapillary assays to assess the functional significance of polar magneto-aerotaxis in the model organism Magnetospirillum gryphiswaldense. We found that a magnetic field configuration matching the predominant swimming polarity of the population results in an up to 4-fold increased peak intensity of the aerotactic band compared to populations with non-matching polarity. Competition assays using fluorescently labeled north- and south-seeking populations confirmed that congruence between swimming polarity and magnetic field orientation markedly improves aerotactic band formation in oxygen gradients. Alongside our main findings, we noted biomagnetism-independent phototactic responses integrated with aerotaxis, driving collective unidirectional migration along the oxygen gradient. Our results provide direct evidence that matching swimming polarity with the magnetic field confers a clear advantage in navigating oxygen gradients. These findings reinforce the role of the geomagnetic field in shaping MTB behavior and highlight the adaptive value of magnetotactic swimming polarity in environmental navigation.

|
Scooped by
mhryu@live.com
Today, 3:58 PM
|
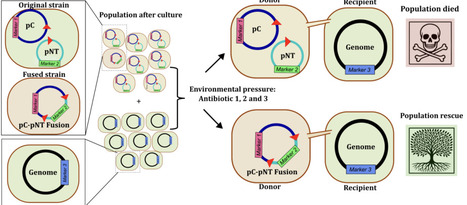
Plasmids are key drivers of bacterial adaptation, yet the mechanisms that generate their diversity remain poorly understood. Here, we show that mobile genetic elements (MGEs) orchestrate a fusion-deletion life cycle that drives the emergence of multireplicon, multidrug-resistant plasmids in Staphylococcus aureus. Large-scale genomic analyses reveal that multireplicon plasmids are widespread and strongly enriched in transposases. Using experimental assays, we demonstrate that rare MGE-mediated fusion events, via homologous recombination or transposition, combine distinct plasmids into single multireplicon elements, expanding gene content and transfer potential. Antibiotic pressure selectively enriches these fused plasmids, rescuing bacterial populations under stress, whereas opposing selective forces, including phage predation, favor deletion derivatives that preserve essential functions and transmissibility. This cyclical process generates dynamic plasmid repertoires with conserved backbones and diverse accessory modules. We propose that MGE-driven fusion-deletion cycles represent a general principle of plasmid evolution, explaining the rapid emergence and persistence of multidrug-resistant plasmids across bacterial pathogens.

|
Scooped by
mhryu@live.com
Today, 3:41 PM
|
Aptamers are highly selective nucleic acid ligands that overcome many challenges of antibodies, including structural instability and high production costs. The implementation of aptamers into routine research and commercial applications has advanced steadily, though it has been tempered by the limitations of existing discovery methods, which remain relatively slow and low-throughput. SELEX, the gold-standard method for aptamer discovery, has played a pivotal role in the field. As the demand for faster and more tailored solutions grows, there is increasing recognition of the value of newer approaches that can expedite the discovery and optimization of aptamers for specific applications. This review discusses recent advances in the methods used for aptamer engineering, including both wet lab methods performed in vitro, and dry lab methods performed in silico. Applications of engineered aptamers described in recent literature and discuss new developments that could further lead to innovative new applications of aptamers for use both within and outside of the laboratory are discussed. Within the laboratory, these applications include (but are not limited to) target-binding assays and integration into analytical nanomaterials, and outside of the laboratory, diagnostics and biosensing, which will be discussed at length.

|
Scooped by
mhryu@live.com
Today, 3:19 PM
|
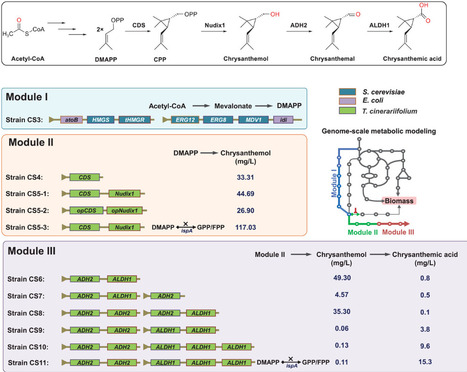
Chrysanthemic acid is an unconventional monoterpene moiety of the natural pesticide pyrethrins with notable anti-insect activity, making its industrial biosynthesis a promising avenue for sustainable agriculture. Here, an E. coli cell factory is designed and build for highly efficient chrysanthemic acid production guided by Genome-scale metabolic models (GEM). The biosynthetic pathway is reconstructed and simulated the metabolic changes caused by exogenous modules are simulated. A key metabolic branch point catalyzed by ispA is identified by this model, and inhibiting its expression using synthetic small RNA redirected the metabolic flux, resulting in the titers of precursor chrysanthemol and chrysanthemic acid increasing by 162% and 59%, respectively. The effect of the expression level of downstream dehydrogenases on chrysanthemic acid titer is also predicated using GEM, and the further optimization of copy number for dehydrogenase genes led to a notably 570% increase in chrysanthemic acid titer experimentally. By integrating the debranching strategy with copy number optimization, a record chrysanthemic acid titer 141.78 mg L−1 is achieved in a bioreactor. The work seamlessly integrated in silico modeling optimization with wet-lab practices that significantly enhance target metabolite titer through metabolic network engineering, offering a new route for constructing efficient cell factories for natural bioproducts.

|
Scooped by
mhryu@live.com
Today, 3:02 PM
|
Respiratory burst oxidase homolog D (RBOHD)-dependent reactive oxygen species (ROS) in Arabidopsis are well known to suppress pathogen colonization, but their influence on beneficial microbes remains unclear. Here, we found that the beneficial rhizobacterium Pseudomonas anguilliseptica was significantly less enriched in the rhizosphere of rbohD mutants than in that of wild-type plants. Conversely, elevated rhizosphere ROS levels, either triggered by pretreatment with pathogenic Dickeya solani bacteria or caused by mutations in ROS scavenging genes (e.g., in apx1 and cat2 mutants), promoted the rhizosphere recruitment of P. anguilliseptica. This promoting effect was abolished by catalase treatment. In situ microfluidic chemotaxis assays further revealed that P. anguilliseptica exhibits a chemotactic response to low concentrations of hydrogen peroxide ( ≤ 500 nM), accompanied by upregulated expression of chemotaxis- and motility-related genes. Notably, inoculation of P. anguilliseptica effectively suppressed D. solani-induced disease symptoms, and this protective effect was attenuated by catalase treatment. Collectively, these findings reveal a previously unrecognized role of ROS in recruitment beneficial microbiota to enhance plant growth and suppress disease symptoms.

|
Scooped by
mhryu@live.com
Today, 2:13 PM
|
The apoplast is an important battlefield in plant–pathogen interactions. The late blight oomycete pathogen Phytophthora infestans, for instance, secretes cystatin-like protease inhibitors EpiC1 and EpiC2B to suppress C14, a papain-like immune protease secreted by tomato. Here, we found that P. infestans also secretes two distinct papain-like proteases termed Pain1 and Pain2, which are transcriptionally induced during infection. Both Pains promote P. infestans infection, but not when their catalytic residues are mutated. Strikingly, EpiC1 and EpiC2B preferentially inhibit tomato C14 rather than self-produced Pains, suggesting that they coevolved with Pains to avoid self-inhibition. To mimic the avoidance of inhibition by EpiCs, we engineered C14 (eC14) with seven Pain1 residues that potentially disturb the EpiCs–C14 interface. This eC14 is less sensitive to inhibition by EpiCs and enhances resistance to P. infestans infection. This strategy demonstrates that a pathogen-inspired protein engineering approach can increase crop resistance to plant pathogens.

|
Scooped by
mhryu@live.com
Today, 1:24 PM
|
Rapid and precise detection of small-molecule metabolites is crucial for optimizing the bioproduction processes. Cell-free systems (CFSs) offer an ideal platform for developing such biosensors due to their speed and suitability for automation. However, transcription factor (TF)-based biosensors, which are key elements for metabolite sensing, suffer from a severe bottleneck in in vitro environments. Their performance is often compromised due to the absence of nucleoid-associated proteins and different DNA topology compared to in vivo conditions. Here, we present a systematic framework for rationally engineering high-performance TF biosensors optimized for CFSs through integrated control of TF availability and promoter sequence. Using an itaconate (ITA)-responsive biosensor regulated by the LysR-type transcriptional regulator ItcR as a model system, we demonstrate that modulating TF supply and redesigning promoter elements substantially enhance sensitivity and dynamic range. Promoter dissection revealed that upstream sequences that function normally in vivo interfered with regulated transcription in CFSs, and that truncation to remove this region restored inducible behavior. Subsequent fine-tuning of the −35 and −10 motifs enhanced RNA polymerase recruitment and regulator interaction, resulting in 19-fold higher maximum signal, a 3.3-fold lower detection limit (0.003 g/L ITA), and a steeper dose-response curve (Hill slope increase from 2.7 to 34.7). The same promoter engineering strategy also improved a 3-hydroxypropionate-responsive biosensor, demonstrating its generality across distinct TF-promoter systems. Collectively, this framework establishes a rational, modular approach for constructing high-performance, topology-aware biosensors in CFSs, directly enabling high-throughput screening and automated biofoundry integration for synthetic biology and metabolic engineering applications.

|
Scooped by
mhryu@live.com
Today, 1:12 PM
|
The presence or absence of some genes in a genome can influence whether other genes are likely to be present or absent. Understanding these gene co-occurrence and avoidance patterns reveals fundamental principles of genome organization, with applications ranging from evolutionary reconstruction to rational design of synthetic genomes. PanForest, presented here, uses random forest classifiers to predict the presence and absence of genes in genomes from the set of other genes present. Performance statistics output by PanForest reveal how predictable each gene’s presence or absence is, based on the presence or absence of other genes in the genome. Further, PanForest produces statistics indicating the importance of each gene in predicting the presence or absence of each other gene. The PanForest software can run serially or in parallel, thereby facilitating the analysis of pangenomes at Network of Life scale. A pangenome of 12,741 accessory genes in 1,000 Escherichia coli genomes was analyzed in around 5 hours using 8 processors. To demonstrate PanForest’s utility, we present a case study and show that certain genes associated with resistance to antimicrobial drugs reliably predict the presence or absence of other genes associated with resistance to the same drug. Further, we highlight several associations between those genes and others not known to be associated with antimicrobial resistance (AMR), or associated with resistance to other drugs. We envisage PanForest’s use in studies from multiple disciplines concerning the dynamics of gene distributions in pangenomes ranging from biomedical science and synthetic biology to molecular ecology.

|
Scooped by
mhryu@live.com
Today, 12:44 PM
|
The growing imperative to ensure food safety, preserve ecological integrity, and mitigate public health risks associated with pesticide residues has driven a critical demand for highly sensitive optical sensors. In this regard, optical biosensors, including fluorescence (FL), colorimetry (CL), surface-enhanced Raman scattering (SERS), surface plasmon resonance (SPR), and chemiluminescence strategies, have been developed for pesticide detection. This review aims to provide a comprehensive summary of both fundamental knowledge and advancements in the field of optical biosensors for pesticide detection. The advantages of these biosensors are highlighted, such as excellent sensitivity, high specificity, and on-site application. Subsequently, a detailed overview of the sensing mechanism of optical biosensors based on different molecular recognition elements [e.g., enzymes, antibodies, aptamers, molecularly imprinted polymers (MIPs), and supramolecular host–guest complexes] is provided. Finally, perspectives are offered on the current challenges and future directions of pesticide biosensors. This review is expected to serve as a fundamental guide for researchers in the field of optical biosensors for pesticide detection and to provide insights and avenues to enhance the performance of existing sensing mechanisms in applications across diverse fields.

|
Scooped by
mhryu@live.com
January 9, 4:55 PM
|
Transcription factors regulate gene expression with DNA-binding domains (DBDs) and activation domains. Despite evidence to the contrary, DBDs are often assumed to be the primary mediators of transcription factor (TF) interactions with DNA and chromatin. Here, we used fast single-molecule tracking of transcription factors in living cells to show that short activation domains can control the fraction of molecules bound to chromatin. Stronger activation domains have higher bound fractions and longer residence times on chromatin. Furthermore, mutations that increase activation domain strength also increase chromatin binding. This trend was consistent in four different activation domains and their mutants. This effect further held for activation domains appended to three different structural classes of DBDs. Stronger activation domains with high chromatin-bound fractions also exhibited increased binding to the p300 coactivator in proximity-assisted photoactivation experiments. Genome-wide measurements indicate these activation domains primarily control the occupancy of binding rather than the genomic location. Taken together, these results demonstrate that very short activation domains play a major role in tethering transcription factors to chromatin.

|
Scooped by
mhryu@live.com
January 9, 4:34 PM
|
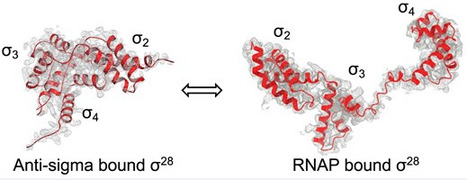
Late flagellar genes in Pseudomonas aeruginosa are transcribed by the group 3 sigma factor, FliA (σ28). σ28 drives the expression of flagellin, which assembles into the flagellar filament in this monoflagellated bacterium. This function is suppressed by the anti-sigma factor FlgM. Here, we present the 1.95Å resolution crystal structure of σ28–FlgM complex, along with a 3.4Å structure of σ28RNAP open promoter complex determined using single particle cryo-electron microscopy from P. aeruginosa. The σ28 adopts a compact conformation upon binding to the anti-sigma factor FlgM, which contacts all three domains of the sigma factor. This conformation is neither conducive to interactions with RNA polymerase nor the promoter DNA. The cryo-EM structure reveals base-specific interactions of σ28 domain 4 (σ4) with −35 element, flipping of −11 base of the template strand, novel interactions of template strand with domain 2 (σ2) and 3 (σ3), and partial insertion of sigma finger into the active site cleft, offering unique features of group 3 sigma interactions with promoter DNA. Perturbation of key residues affects transcription in vitro and flagellar phenotypes as well as bacterial motility in vivo. Analysis of the structural data presented here reveals new insights into transcription regulation of late flagellar genes.
|

|
Scooped by
mhryu@live.com
Today, 4:43 PM
|
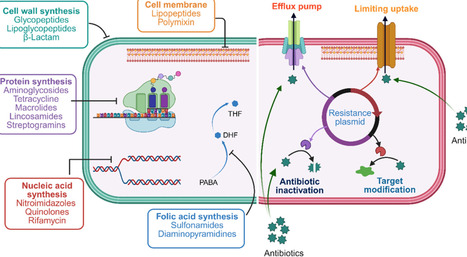
Antibiotics have revolutionized human health by significantly reducing morbidity and mortality associated with bacterial infections. Antibiotics exert bactericidal or bacteriostatic effects through inhibition of cell wall synthesis and disruption of cell membrane integrity, inhibition of protein, nucleic acid synthesis, and other metabolic pathways. Despite their remarkable success since the mid-20th century, antimicrobial resistance (AMR) has emerged as a major global health concern, undermining current treatments and complicating infection management. Key drivers of AMR include the overuse and misuse of antibiotics in clinical settings as well as bacterial adaptations such as genetic mutations and horizontal gene transfer. Mechanistically, these changes can lead to enzymatic inactivation of antibiotics, modification of drug targets, changes in permeability, and active efflux of antimicrobial agents. As resistance rises, antibiotic discovery and development have lagged, creating an urgent need for novel therapeutic strategies and chemical scaffolds. This review examines the antibiotic mechanisms and antibiotic evasion strategies, highlighting genetic and omics approaches used to identify high-priority targets for future drug discovery.

|
Scooped by
mhryu@live.com
Today, 4:37 PM
|
Bacteria survive hostile conditions in clinically relevant conditions by shutting down protein synthesis, but how they restart growth remains poorly understood. Here, we use an E. coli delta-rimM strain, which exhibits a prolonged growth arrest, as a model to investigate how bacteria recover from this arrested state and restore protein synthesis. RimM is a conserved ribosome maturation factor for the 3'-major (head) domain of the 16S rRNA within the bacterial 30S subunit. The loss of RimM causes a significantly longer delay in recovery than other 30S maturation factors, including RbfA - the presumed primary factor in 30S maturation. Cryo-EM analysis of delta-rimM ribosomes revealed a delayed recruitment of ribosomal proteins to the 30S head domain and increased occupancy of the initiation factors IF1 and IF3, as well as recruitment of the silencing factor RsfS to the 50S subunit. These coordinated changes provide a safeguarding mechanism to block the assembly of premature 70S ribosomes. Notably, while the delayed 30S assembly in delta-rimM reduces the activity of global protein synthesis during the recovery phase, bacteria attempt to compensate for this deficiency by producing higher levels of the ribosomal machinery, indicating a programmatic change in energy allocation to generate the ribosome machinery. These findings highlight the importance of the RimM-assisted assembly of the ribosomal head domain for bacterial recovery from growth arrest.

|
Scooped by
mhryu@live.com
Today, 4:24 PM
|
Microbial communities often rely on metabolic cooperation, especially under nutrient limitation, but how antibiotics influence these interactions remains unclear. Here, we show that exposure to chloramphenicol, a ribosome-targeting bacteriostatic antibiotic, promotes cooperation in auxotrophic Escherichia coli consortia. Using a proteome-partition model, we find that chloramphenicol-induced growth inhibition elevates levels of the alarmone ppGpp, shifting proteome allocation toward amino acid biosynthesis and export. This regulatory response enhances cross-feeding and supports cooperative growth, even under antibiotic stress. Laboratory experiments confirm that, under low amino acid availability, co-cultures of leucine and phenylalanine auxotrophs display greater tolerance to chloramphenicol than monocultures. Both our theoretical and experimental results reveal a feedback loop in which antibiotic stress strengthens metabolic interdependence, buffering growth inhibition and enhancing community-level tolerance.

|
Scooped by
mhryu@live.com
Today, 3:57 PM
|
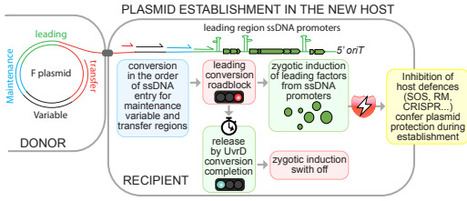
The rapid global spread of antibiotic resistance is mediated by conjugative plasmids, yet how these elements establish immediately after entering new bacterial hosts remains poorly understood. Here we show that plasmids actively coordinate DNA processing with early gene expression to promote their own establishment. Using single-cell measurements of plasmid conversion dynamics, we demonstrate that the F plasmid selectively delays complementary strand synthesis within its leading region, creating a transient single-stranded DNA window that prolongs zygotic induction. This delay is imposed by a single-stranded DNA promoter that forms a structural roadblock to complementary strand DNA synthesis and is resolved by the host helicase UvrD. By extending the single-stranded state of the leading region, plasmids enhance early expression of protective genes that counter host defence responses and promote survival during entry, particularly in non-isogenic hosts. Together, these findings identify early post-entry regulation as a critical determinant of plasmid establishment and reveal how coupling DNA conversion to transient protective gene expression enables conjugative plasmids to overcome host barriers and disseminate antibiotic resistance.

|
Scooped by
mhryu@live.com
Today, 3:26 PM
|
In recent years, DNA origami technology has advanced rapidly as a groundbreaking method for nanomanufacturing. This technology takes advantage of the unique base-pairing characteristics of DNA, and has significant advantages in constructing spatially ordered and programmable nanostructures. This capability aligns with synthetic biology's core principle of mimicking, extending, and reconstructing natural biological processes by modularly assembling artificial systems. This article provides a comprehensive overview of DNA origami's innovative applications across various domains, including cell membrane surfaces, intercellular communication, intelligent biosensing, and precise gene editing, progressing from the extracellular to the intracellular environment. Finally, this review highlights the synergistic interaction between this technology and cell-free synthetic biology, achieved through the integration of in vitro assembly and cellular regulation, thereby opening new pathways for the rational design of artificial life systems.

|
Scooped by
mhryu@live.com
Today, 3:15 PM
|
Inspired by the natural ability of bacteriophages to deliver genetic material directly into host cells, we employed a bottom-up approach to construct a multifunctional synthetic DNA origami needle-like structure. This origami is functionalized with trastuzumab antibodies, cholesterol, protective polymers, and two dyes, which together enable selective targeting and insertion into SKBR3 cancer cells. A disulfide-linked dye payload was attached to the apex of the needle, allowing controlled release in the cytoplasm triggered by the high intracellular glutathione concentration. Real-time tracking of the payload confirmed both successful targeting of the origami structure and subsequent direct cytosolic delivery. By mimicking fundamental mechanisms of bacteriophages, we propose that this artificial needle structure can serve as a prototypical device for the targeted delivery of small-molecule drugs directly into the cytosol.

|
Scooped by
mhryu@live.com
Today, 2:22 PM
|
Adenosine-to-inosine (A-to-I) RNA editing by ADAR enzymes shapes transcript fate and underpins emerging RNA editing therapeutics, yet predicting which adenosines are edited remains difficult. We introduce ADAR-GPT, a model-agnostic fine-tuning framework that adapts a GPT-class language model to classify editing at candidate sites using sequence context in standardized 201 nt windows with the target adenosine explicitly marked. We train and evaluate on GTEx liver data (n=131 samples) at a clinically relevant 15% editing threshold, using a two-stage continual fine-tuning approach where lower thresholds serve as curriculum data to progressively sharpen decision boundaries. Using sequence data, ADAR-GPT demonstrates competitive or superior performance when benchmarked against established computational approaches, including convolutional and foundation model architectures, achieving a better balance of recall, precision, and specificity alongside stronger operating-curve metrics. The approach is reproducible and portable across GPT backbones without architectural changes. Beyond accurate site classification, ADAR-GPT provides practical adenosine scoring to prioritize experimental targets and inform guide RNA design, with a framework adaptable to new datasets and model architectures.

|
Scooped by
mhryu@live.com
Today, 1:40 PM
|
Microbes play a pivotal role in the Earth’s carbon cycle, regulating greenhouse gas fluxes by emitting, fixing and transforming CO2. Among them, acetogens stand out for their ability to fix CO2 through the Wood–Ljungdahl pathway, an ancient, highly energy-efficient route to acetyl-CoA that operates at thermodynamic limits. By coupling hydrogen (H2) or carbon monoxide oxidation to CO2 fixation, acetogens conserve energy while generating biomass and valuable products such as ethanol and acetate. These features position them as promising microbial cell factories for sustainable bioproduction via gas fermentation. Recent advances in metabolic engineering and synthetic biology have expanded the production spectrum of acetogens, enabling production of platform chemicals at lab-to-commercial scale. Yet, CO2-only bioconversion remains energetically challenging compared to syngas-based applications, requiring innovative solutions in strain development, bioprocess optimisation and integration of renewable energy sources. This review highlights the central role of model acetogens in anaerobic CO2 conversion, covering their metabolic capabilities, strain development and emerging bioprocess strategies to unlock their potential for low-carbon biomanufacturing.

|
Scooped by
mhryu@live.com
Today, 1:20 PM
|
Designing proteins that bind with high affinity to hydrophilic protein target sites remains a challenging problem. Here we show that RFdiffusion can be conditioned to generate protein scaffolds that form geometrically matched extended β-sheets with target protein edge β-strands in which polar groups on the target are complemented with hydrogen bonding groups on the design. We use this approach to design binders against edge-strand target sites on KIT, PDGFRɑ, ALK-2, ALK-3, FCRL5, NRP1, and α-CTX, and obtain higher (pM to mid nM) affinities and success rates than unconditioned RFdiffusion. Despite sharing β-strand interactions, designs have high specificity, reflecting the precise customization of interacting β-strand geometry and additional designed binder-target interactions. A binder-KIT co-crystal structure is nearly identical to the design model, confirming the accuracy of the design approach. The ability to robustly generate binders to the hydrophilic interaction surfaces of exposed β-strands considerably increases the range of computational binder design. This study demonstrates the capability of deep learning protein design models in generating functionally validated β-strand pairing interfaces, expanding the structural diversity of de novo binding proteins and accessible target surfaces.

|
Scooped by
mhryu@live.com
Today, 12:50 PM
|
The rapid growth of biodiesel production generates large amounts of crude glycerol, a low-value byproduct with environmental and economic challenges. Here, we present an engineered yeast coculture system combining Saccharomyces cerevisiae and Yarrowia lipolytica to convert crude glycerol into isopropanol, a liquid organic hydrogen carrier. The system integrates metabolic engineering, cell surface display pairing, immobilization, and continuous cultivation in fibrous bed bioreactors. In S. cerevisiae, glycerol use was improved by transporter optimization, pathway redirection, and flux shift from ethanol to isopropanol. In Y. lipolytica, ethanol from S. cerevisiae was redirected to isopropanol by acetyl-CoA reinforcement, malonyl-CoA diversion, and NADPH availability. Optimized pairing and inoculation ratios enhanced stability and yield. The consortia achieved complete glycerol utilization and three reuse cycles over 180 h. With pure glycerol, 28.34 g/l isopropanol was produced, while crude glycerol reached 86.06% of this yield. This strategy offers a scalable, modular route to convert biodiesel byproducts into bioenergy carriers.

|
Scooped by
mhryu@live.com
January 9, 5:06 PM
|
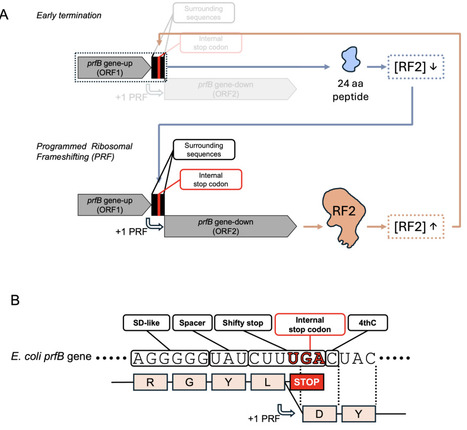
Understanding why some traits are maintained whereas others are repeatedly lost is a central question in evolutionary biology. Here, we address this problem through an analysis of the evolutionary dynamics of autoregulation of the prfB, which encodes peptide-chain release factor 2 (RF2), a key factor in bacterial translation termination. RF2 recognizes UGA and UAA stop codons and catalyzes the release of the completed polypeptide. In many species, prfB contains an internal UGA stop codon, triggering premature translation termination by RF2 itself. Complete RF2 translation depends on a +1 programmed ribosomal frameshifting (PRF) event on the internal stop codon, which occurs more frequently when RF2 levels are low, resulting in autoregulation of prfB expression. While widespread, this autoregulatory mechanism has been lost in multiple bacterial lineages. We combined phylogenetics, experimental evolution and molecular genetics to investigate the evolutionary forces behind this loss. We found no significant correlation between PRF loss and stop codon usage using phylogenetically informed analyses, and PRF disruption in Pseudomonas fluorescens SBW25 had no detectable fitness effect. However, engineered mutations that reduced frameshifting caused fitness defects, which were compensated by two classes of mutation: (i) mutations that impair specific ribosomal proteins, and (ii) single-nucleotide deletions in prfB that adjust the reading frame and bypass the internal stop codon. These results suggest that compensatory mutations facilitate the loss of prfB autoregulation under RF2-limiting conditions. We discuss three potential scenarios that could account for this process.

|
Scooped by
mhryu@live.com
January 9, 4:45 PM
|
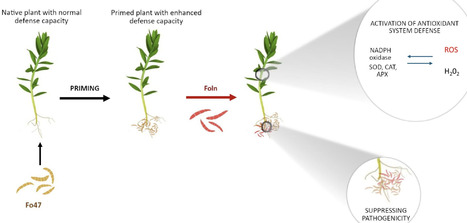
Plants rely on specialized adaptive mechanisms to enhance resistance against environmental stress. One such mechanism, priming, enables faster and stronger defence responses upon subsequent stress exposure. This study examines whether the non-pathogenic Fusarium oxysporum Fo47 primes flax by colonizing roots and activating antioxidant defences. Flax plants primed with Fo47 and those treated with both Fo47 and the pathogenic strain F. oxysporum Foln were analysed for fungal colonization, PR genes expression and antioxidant systems: enzymatic (ROS metabolism-related genes expression, catalase and superoxide dismutase activity, hydrogen peroxide and superoxide anion levels) and non-enzymatic (phenolic compound content and antioxidant potential). The results demonstrate that Fo47 colonises host tissues, significantly reducing Foln penetration and colonisation, particularly in primed plants. Root-specific suppression of Foln by Fo47 was stronger than systemic suppression in shoots. Fo47 induced early chitinase and NADPH oxidases D transcript accumulation and reduced superoxide anion level in roots, likely triggering defence activation. Notably, Fo47 also activated both enzymatic and non-enzymatic antioxidant systems in shoots, suggesting a systemic priming effect. These findings underscore the potential of non-pathogenic F. oxysporum strains in sustainable plant protection strategies.
|
 Your new post is loading...
Your new post is loading...




























lee sy, 2st