 Your new post is loading...

|
Scooped by
?
May 2, 5:17 PM
|
Genetic variance is vital for breeding programs and mutant screening, yet traditional mutagenesis methods wrestle with genetic redundancy and a lack of specificity in gene targeting. CRISPR-Cas9 offers precise, site-specific gene editing, but its application in crop improvement has been limited by scalability challenges. In this study, we develop genome-wide multi-targeted CRISPR libraries in tomato, enhancing the scalability of CRISPR gene editing in crops and addressing the challenges of redundancy while maintaining its precision. We design 15,804 unique single guide RNAs (sgRNAs), each targeting multiple genes within the same gene families. These sgRNAs are classified into 10 sub-libraries based on gene function. We generate approximately 1300 independent CRISPR lines and successfully identify mutants with distinct phenotypes related to fruit development, fruit flavor, nutrient uptake, and pathogen response. Additionally, we develop CRISPR-GuideMap, a double-barcode tagging system to enable large-scale sgRNA tracking in generated plants. Our results demonstrate that multi-targeted CRISPR libraries are scalable and effective for large-scale gene editing and offer an approach to overcome gene functional redundancy in basic plant research and crop breeding. Genetic variance is vital for breeding programs and mutant screening, yet traditional mutagenesis methods wrestle with genetic redundancy and a lack of specificity in gene targeting. Here the authors develop a scalable CRISPR system in tomato to edit multiple genes at once, overcoming genetic redundancy and revealing new traits related to fruit quality, nutrient use, and disease resistance.

|
Scooped by
?
May 2, 4:31 PM
|
The application of beneficial, plant-associated microorganisms is a sustainable approach to improving crop performance in agriculture. However, microbial inoculants are often susceptible to prolonged periods of storage and deleterious environmental factors, which negatively impact their viability and ultimately limit efficacy in the field. This particularly concerns non-sporulating bacteria. To overcome this challenge, the availability of protective formulations is crucial. Numerous parameters influence the viability of microbial cells, with drying procedures generally being among the most critical ones. Thus, technological advances to attenuate the desiccation stress imposed on living cells are key to successful formulation development. In this review, we discuss the core aspects important to consider when aiming at high cell viability of non-sporulating bacteria to be applied as microbial inoculants in agriculture. We elaborate the suitability of commonly applied drying methods (freeze-drying, vacuum-drying, spray-drying, fluidized bed-drying, air-drying) and potential measures to prevent cell damage from desiccation (externally applied protectants, stress pre-conditioning, triggering of exopolysaccharide secretion, ‘helper’ strains). Furthermore, we point out methods for assessing bacterial viability, such as colony counting, spectrophotometry, microcalorimetry, flow cytometry and viability qPCR. Choosing appropriate technologies for maintenance of cell viability and evaluation thereof will render formulation development more efficient. This in turn will aid in utilizing the vast potential of promising, plant beneficial bacteria as sustainable alternatives to standard agrochemicals.

|
Scooped by
?
May 2, 2:14 AM
|
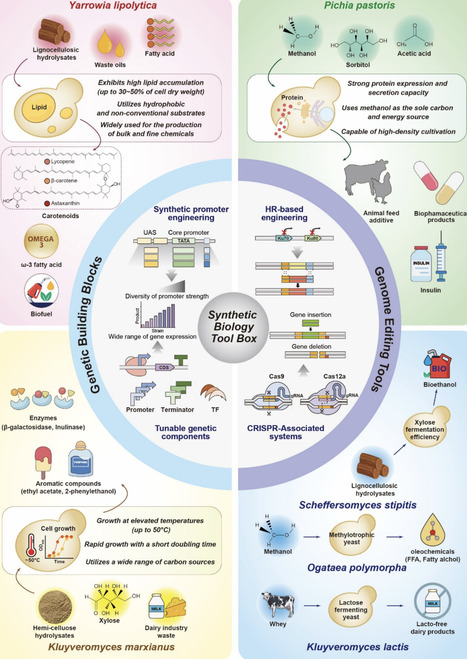
Non-conventional yeasts exhibit exceptional genetic and functional diversity, serving as a largely untapped repertoire for biotechnological applications. Beyond the conventional yeast Saccharomyces cerevisiae, non-conventional yeasts are naturally more multifaceted, possessing the ability to utilize renewable and low-cost carbon sources while exhibiting robust physiology under challenging conditions. However, their vast potential remains largely unexplored, encompassing both challenges and opportunities for biotechnological advancements. Over the past decade, technological advancements in synthetic biology have unlocked new opportunities to harness their potential and overcome inherent limitations, enabling the full exploitation of their advantages across a broad spectrum of applications. In this review, we highlight recent advances in the synthetic biology of non-conventional yeasts, focusing on the development of new genetic building blocks (e.g., promoters and terminators), genome editing tools, and metabolic pathway engineering. Through these technologies, non-conventional yeasts are poised to emerge as pivotal next-generation workhorses tailored for specific applications in sustainable biomanufacturing, accelerating the transition to a bio-based economy.

|
Scooped by
?
May 2, 1:48 AM
|
The complex and mutual interactions between plants and their associated microbiota are key for plant survival and fitness. From the myriad of microbes that exist in the soil, plants dynamically engineer their surrounding microbiome in response to varying environmental and nutrient conditions. The notion that the rhizosphere bacterial and fungal community acts in harmony with plants is widely acknowledged, yet little is known about how these microorganisms interact with each other and their host plants. Here, we explored the interaction of two well-studied plant beneficial endophytes, Enterobacter sp. SA187 and the fungus Serendipita indica. We show that these microbes show inhibitory growth in vitro but act in a mutually positive manner in the presence of Arabidopsis as a plant host. Although both microbes can promote plant salinity tolerance, plant resilience is enhanced in the ternary interaction, revealing that the host plant has the ability to positively orchestrate the interactions between microbes to everyone's benefit. In conclusion, this study advances our understanding of plant–microbiome interaction beyond individual plant–microbe relationships, unveiling a new layer of complexity in how plants manage microbial communities for optimal growth and stress resistance.

|
Scooped by
?
May 2, 1:34 AM
|
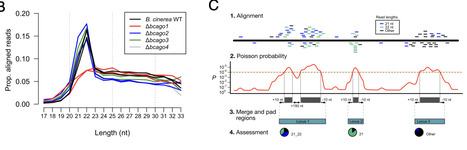
Argonaute (AGO) proteins bind to small RNAs to induce RNA interference (RNAi), a conserved gene regulatory mechanism in animal, plant, and fungal kingdoms. Small RNAs of the fungal plant pathogen Botrytis cinerea were previously shown to translocate into plant cells and to bind to the host AGO, which induced cross-kingdom RNAi to promote infection. However, the role of pathogen AGOs during host infection stayed elusive. In this study, we revealed that members of fungal plant pathogen B. cinerea BcAGO family contribute to plant infection. BcAGO1 binds to both fungal and plant small RNAs during infection and acts in bidirectional cross-kingdom RNAi, from fungus to plant and vice versa. BcAGO2 also binds fungal and plant small RNAs but acts independent from BcAGO1 by regulating distinct genes. Nevertheless, BcAGO2 is important for infection, as it is required for effective pathogen small RNA delivery into host cells and fungal induced cross-kingdom RNAi. Providing these mechanistic insights of pathogen AGOs promises to improve RNAi-based crop protection strategies.

|
Scooped by
?
May 2, 12:56 AM
|
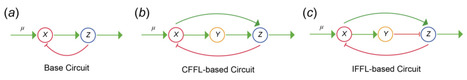
Perfect adaptation, the ability to regulate and maintain gene expression to its desired value despite disturbances, is important in the development of organisms. Building biological controllers to endow engineered biological systems with such perfect adaptation capability is a key goal in synthetic biology. Model-guided exploration of such synthetic circuits has been effective in designing such systems. However, theoretical analysis to guarantee controller properties with nonlinear models, such as Hill functions, remains challenging, while use of linear models fails to capture the inherent nonlinear dynamics of gene expression systems. Here, we propose a reverse engineering approach to infer the kinetic parameters for nonlinear Hill function-type models from analysis of linear models and apply our method to design controllers, which achieve perfect adaptation. Focusing on three biological network motif-based controllers, we demonstrate via simulation the efficacy of the proposed approach in combining linear system theories with nonlinear modelling, to design multiple gene circuits that could deliver perfect adaptation. Given the ubiquitous use of Hill functions in describing the dynamics of biological regulatory networks, we anticipate the proposed reverse engineering approach to benefit a wide range of systems and synthetic biology applications.

|
Scooped by
?
May 2, 12:36 AM
|
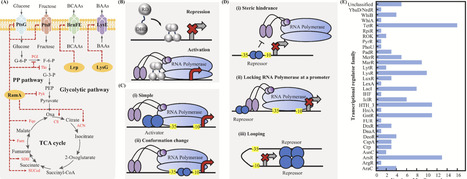
Intracellular biosensors based on transcriptional regulators have become essential instruments in biomanufacturing, extensively employed for the semi-quantitative assessment of intracellular metabolites, high-throughput screening of production strains, and the directed evolution of enzymes. Corynebacterium glutamicum serves as an industrial chassis for the production of amino acids and a variety of high-value-added chemicals. This paper discusses the varieties and modes of action of transcriptional regulators employed in the construction of intracellular biosensors in C. glutamicum. It also reviews the design principles and progress in the application of transcriptional regulator-based biosensors. Furthermore, measures designed to improve the efficacy of these biosensors are delineated. The challenges and future prospects of biosensors based on transcriptional regulators in practical applications are analyzed. This review seeks to offer theoretical direction for the systematic design and development of transcriptional regulator-based biosensors and to aid researchers in enhancing the growth and productivity of microbial production strains.

|
Scooped by
?
May 2, 12:26 AM
|
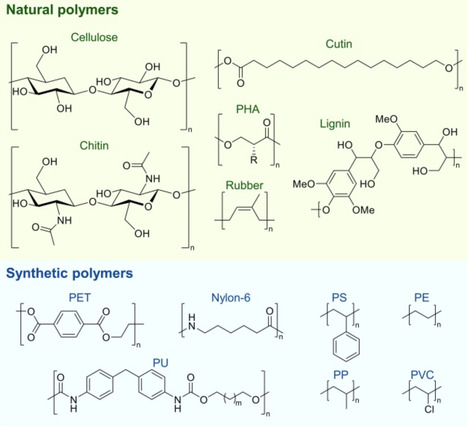
Since the 1950s, plastics have become commodity materials that are present in virtually every aspect of our daily lives. However, the current economic model of plastics is fundamentally linear, with less than 10% of plastics returning to the value chain at their end of life. In recent years, efforts have been dedicated to develop new technologies that can change this model to a circular economy for plastics, including enzymatic recycling and biological upcycling to value-added products. Here, we will review recent advances made in this rapidly evolving field and discuss how further development of these technologies could contribute to reduce the share of postconsumer plastic waste that is diverted toward landfilling and incineration.

|
Scooped by
?
May 2, 12:16 AM
|
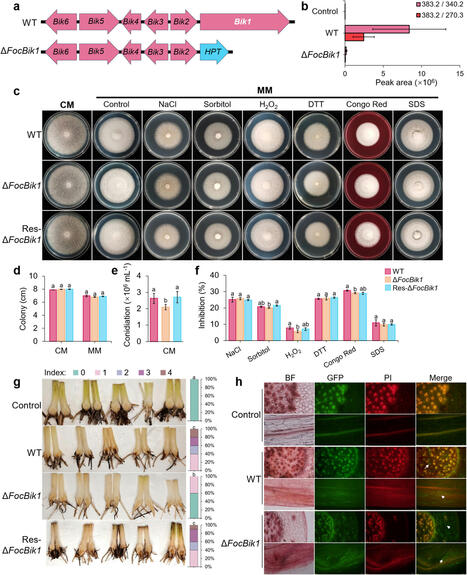
Fusarium wilt, caused by Fusarium oxysporum f. sp. cubense Tropical Race 4 (Foc TR4), poses a severe threat to global banana production. Secondary metabolites are critical tools employed by pathogens to interact with their environment and modulate host–pathogen dynamics. Bikaverin, a red-colored polyketide pigment produced by several Fusarium species, has been studied for its pharmacological properties, but its ecological roles and impact on pathogenicity remain unclear. This study investigated the role of bikaverin in Foc TR4, focusing on its contribution to pathogenicity and its interaction with the rhizosphere microbiome. Pathogenicity assays under sterile and autoclaved conditions demonstrated that bikaverin does not directly contribute to pathogenicity by affecting the infection process or damaging host tissues. Instead, bikaverin indirectly enhances Foc TR4’s pathogenicity by reshaping the rhizosphere microbiome. It suppresses beneficial plant growth-promoting rhizobacteria, such as Bacillus, while promoting the dominance of fungal genera, thereby creating a microbial environment beneficial for pathogen colonization and infection. Notably, bikaverin biosynthesis was found to be tightly regulated by environmental cues, including acidic pH, nitrogen scarcity, and microbial competition. Co-culture with microbes such as Bacillus velezensis and Botrytis cinerea strongly induced bikaverin production and upregulated expression of the key bikaverin biosynthetic gene FocBik1. In addition, the identification of bikaverin-resistant Bacillus BR160, a strain with broad-spectrum antifungal activity, highlights its potential as a biocontrol agent for banana wilt management, although its stability and efficiency under field conditions require further validation. Bikaverin plays an indirect yet important role in the pathogenicity of Foc TR4 by manipulating the rhizosphere microbiome. This ecological function underscores its potential as a target for sustainable disease management strategies. Future research should focus on elucidating the molecular mechanisms underlying bikaverin-mediated microbial interactions, using integrated approaches such as transcriptomics and metabolomics. Together, these findings provide a foundation for novel approaches to combat banana wilt disease and enhance crop resistance.

|
Scooped by
?
May 1, 11:46 PM
|
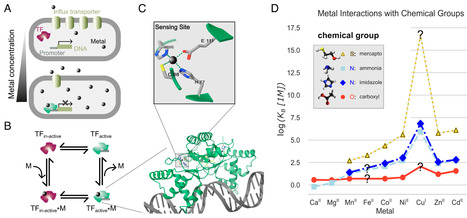
In the common cellular space, hundreds of binding reactions occur reliably and simultaneously without disruptive mutual interference. The design principles that enable this remarkable compatibility have not yet been adequately elucidated. In order to delineate these principles, we consider the intracellular sensing of transition metals in bacteria—an integral part of cellular metal homeostasis. Protein cytosolic sensors typically interact with metals through three types of lateral chain residues, containing oxygen, nitrogen, or sulfur. The very existence of complete sets of mutually compatible sensors is a nontrivial problem solved by evolution, since each metal sensor has to bind to its cognate metal without being “mismetallated” by noncognate competitors. Here, based solely on theoretical considerations and limited information about binding constants for metal-amino acid interactions, we are able to predict possible “sensor compositions,” i.e., the residues forming the binding sites. We find that complete transition-metal sensor sets are severely limited in their number by compatibility requirements, leaving only a handful of possible sensor compositions for each transition metal. Our theoretical results turn out to be broadly consistent with experimental data on known bacterial sensors. If applicable to other cytosolic binding interactions, the results generated by our approach imply that compatibility requirements may play a crucial role in the organization and functioning of intracellular processes.

|
Scooped by
?
May 1, 11:18 PM
|
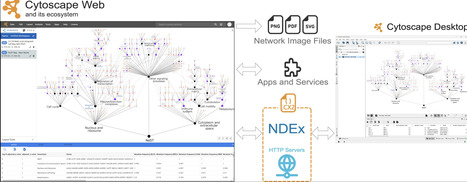
Since its introduction in 2003, Cytoscape has been a de facto standard for visualizing and analyzing biological networks. We now introduce Cytoscape Web (https://web.cytoscape.org), an online implementation that captures the interface and key visualization functionality of the desktop while providing integration with web tools and databases. Cytoscape Web enhances accessibility, simplifying collaboration through online data sharing. It integrates with Cytoscape desktop via the CX2 network exchange format and with the Network Data Exchange for storing and sharing networks. The platform supports extensibility through an App framework for UI components and Service Apps for algorithm integration, fostering community-driven development of new analysis tools. Overall, Cytoscape Web enhances network biology by providing a versatile, accessible, and collaborative online platform that adapts to evolving computational challenges, laying a foundation for future incorporation of advanced network analysis capabilities by the community.

|
Scooped by
?
May 1, 10:50 PM
|
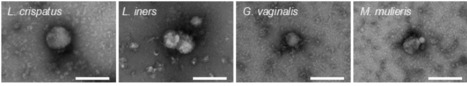
The composition of the vaginal microenvironment has significant implications for gynecologic and obstetric outcomes. Where a Lactobacillus-dominated microenvironment is typically considered optimal, a polymicrobial environment is associated with increased risk for female reproductive diseases. Recent work has examined bacteria-derived extracellular vesicles (bEVs) as an important mode of microbe-host communication in the female reproductive tract, with bEVs exhibiting unique species- and strain-level functions that may influence women's health outcomes. However, in order to communicate with female reproductive tissues, bEVs must be able to penetrate the protective cervicovaginal mucus barrier. As the first line of defense against bacteria and pathogens, cervicovaginal mucus protects against infection in the female reproductive tract through steric, hydrophobic, and electrostatic interactions with foreign pathogens. Here, we hypothesize that the physical properties of bacteria-derived extracellular vesicles enable their mobility through cervicovaginal mucus and permit interactions with upper female reproductive tract tissues. We demonstrate that the barrier properties of mucus allow increased diffusion of bEVs, compared to whole bacteria. We evaluate the uptake of bEVs by, and the resulting effects on, human vaginal epithelial, endometrial, and placental cells, highlighting potential mechanisms of action by which vaginal dysbiosis contributes to gynecologic and obstetric diseases. Our work demonstrates the ability of bEVs to mediate female reproductive diseases and highlights their potential as therapeutic modalities for treating dysbiosis and dysbiosis-associated diseases in the female reproductive tract.

|
Scooped by
?
May 1, 10:05 PM
|
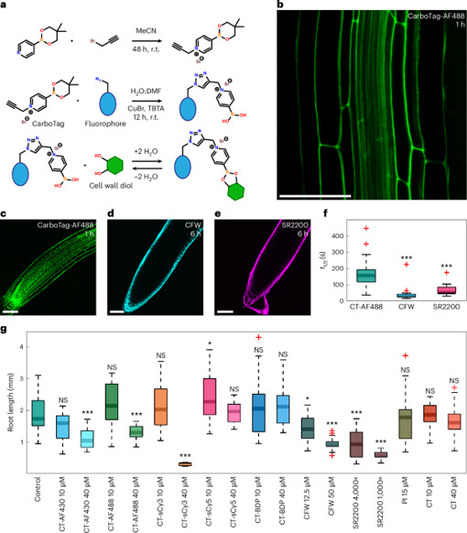
Plant cells are contained within a rigid network of cell walls. Cell walls serve as a structural material and a crucial signaling hub vital to all aspects of the plant life cycle. However, many features of the cell wall remain enigmatic, as it has been challenging to map its functional properties in live plants at subcellular resolution. Here, we introduce CarboTag, a modular toolbox for live functional imaging of plant walls. CarboTag uses a small molecular motif, a pyridine boronic acid, that directs its cargo to the cell wall. We designed a suite of cell wall imaging probes based on CarboTag in various colors for multiplexing. Additionally, we developed new functional reporters for live quantitative imaging of key cell wall characteristics: network porosity, cell wall pH and the presence of reactive oxygen species. CarboTag paves the way for dynamic and quantitative mapping of cell wall responses at subcellular resolution. A suite of nontoxic and permeable CarboTag probes in various colors enables live quantitative imaging of a range of plant cell wall characteristics and dynamic, high-resolution mapping of cell wall changes in response to growth or perturbations.
|

|
Scooped by
?
May 2, 5:14 PM
|
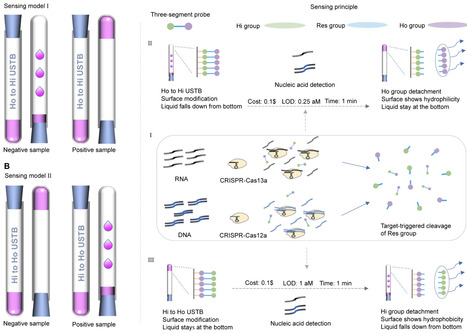
Affordable, sensitive, and simplified DNA/RNA detection is important for disease diagnosis and enables timely medical intervention measures. Usually, high sensitivity depends on expensive instruments and sophisticated procedures, making sensitivity contradict affordability and simplicity. Here, we proposed an ultra-sensitive single-tube biosensor (USTB) where users can visually detect targets by observing the liquid motion state in a glass tube. The developed instrument-free USTB performed low-cost ($0.1), fast (1 min), and ultra-sensitive detection for both the DNA/RNA fragments (≤1 aM) and the clinical positive samples, which commercial reverse transcription polymerase chain reaction (RT-PCR) and PCR kits could not effectively recognize. Furthermore, USTB is promising to be easily applied to detect other-type biomarkers by the designed smart sensing unit.

|
Scooped by
?
May 2, 2:17 AM
|
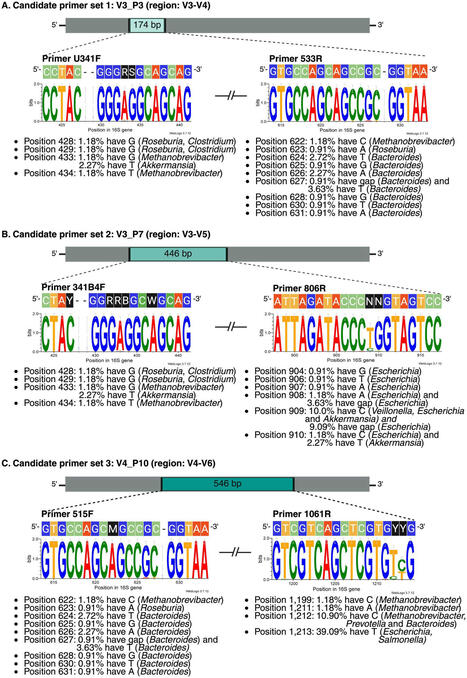
The 16S rRNA gene is crucial for bacterial identification, but primer biases and intergenomic variation can compromise its effectiveness, especially in complex ecosystems like the human gut microbiome. This study systematically evaluates 57 commonly used 16S rRNA primer sets through in silico PCR simulations against the SILVA database. We identified three promising primer sets (V3_P3, V3_P7, and V4_P10) that offer balanced coverage and specificity across 20 key genera of the core gut microbiome. Our findings reveal: (1) significant limitations in widely used “universal” primers, often failing to capture microbial diversity due to unexpected variability in conserved regions, (2) substantial intergenomic variation, even within traditionally conserved regions of the 16S rRNA gene, as demonstrated by Shannon entropy analysis, and (3) discrepancies between intergenomic patterns in NCBI and SILVA databases, highlighting the impact of database choices on taxonomic classification. These results challenge assumptions about 16S rRNA gene conservation and emphasize the need for tailored primer design informed by comprehensive sequence databases. We advocate for a multi-primer strategy to improve coverage and mitigate biases, ultimately enhancing the accuracy and reliability of gut microbiome profiling. This approach has potential applications beyond gut microbiome studies, including animal microbiome research and probiotic community profiling.

|
Scooped by
?
May 2, 2:09 AM
|
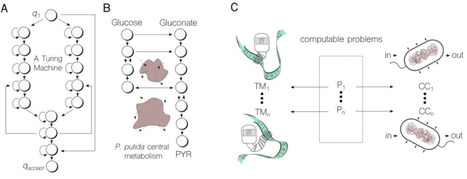
Biological systems inherently perform computations, inspiring synthetic biologists to engineer biological systems capable of executing predefined computational functions for diverse applications. Typically, this involves applying principles from the design of conventional silicon-based computers to create novel biological systems, such as genetic Boolean gates and circuits. However, the natural evolution of biological computation has not adhered to these principles, and this distinction warrants careful consideration. Here, we explore several concepts connecting computational theory, living cells, and computers, which may offer insights into the development of increasingly sophisticated biological computations. While conventional computers approach theoretical limits, solving nearly all problems that are computationally solvable, biological computers have the opportunity to outperform them in specific niches and problem domains. Crucially, biocomputation does not necessarily need to scale to rival or replicate the capabilities of electronic computation. Rather, efforts to re-engineer biology must recognise that life has evolved and optimised itself to solve specific problems using its own principles. Consequently, intelligently designed cellular computations will diverge from traditional computing in both implementation and application.

|
Scooped by
?
May 2, 1:45 AM
|
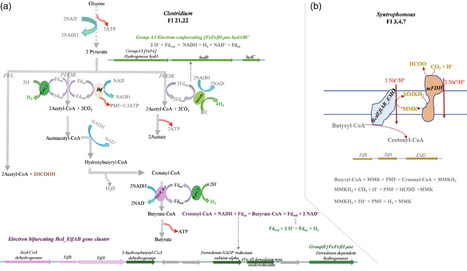
A combined enrichment experiment and genome-centric meta-omics analysis demonstrated that metabolic specificity, rather than flexibility, governs the anaerobic digestion (AD) ecosystem. This study provides new insights into interspecies electron transfer in the AD process, highlighting a segregation in the metabolism of H2 and formate. Our findings show that H2 acts as the primary electron sink for recycling redox cofactors, including NAD+ and oxidised ferredoxin (Fdox), during primary fermentation, while formate is the dominant electron carrier in secondary fermentation, especially under conditions with elevated H2 concentrations. Importantly, no evidence of biochemical interconversion between H2 and formate was identified in the primary fermenting bacteria or in syntrophs enriched in this study. This segregation of H2 and formate metabolism likely benefits the anaerobic oxidation of butyrate and propionate with a higher tolerance to H2 accumulation. Moreover, this study highlights the functional partitioning among microbial populations in key AD niches: primary fermentation, secondary fermentation (syntrophic acetogenesis), hydrogenotrophic methanogenesis, and acetoclastic methanogenesis. Genome-centric analysis of the AD microbiome identified several key functional gene clusters, which could enhance genome-centric genotype–phenotype correlations, particularly for strict anaerobes that are difficult to isolate and characterise in pure culture.

|
Scooped by
?
May 2, 1:13 AM
|
RNAs are critical regulators of gene expression, and their functions are often mediated by complex secondary and tertiary structures. Structured regions in RNA can selectively interact with small molecules—via well-defined ligand-binding pockets—to modulate the regulatory repertoire of an RNA. The broad potential to modulate biological function intentionally via RNA–ligand interactions remains unrealized, however, due to challenges in identifying compact RNA motifs with the ability to bind ligands with good physicochemical properties (often termed drug-like). Here, we devise fpocketR, a computational strategy that accurately detects pockets capable of binding drug-like ligands in RNA structures. Remarkably few, roughly 50, of such pockets have ever been visualized. We experimentally confirmed the ligandability of novel pockets detected with fpocketR using a fragment-based approach introduced here, Frag-MaP, that detects ligand-binding sites in cells. Analysis of pockets detected by fpocketR and validated by Frag-MaP reveals dozens of sites able to bind drug-like ligands, supports a model for RNA secondary structural motifs able to bind quality ligands, and creates a broad framework for understanding the RNA ligand-ome.

|
Scooped by
?
May 2, 12:47 AM
|
Ethanol is a fermentation product widely used as a fuel and chemical precursor in various applications. However, its accumulation imposes severe stress on the microbial producer, leading to significant production losses. To address this, improving a strain’s ethanol tolerance is considered an effective strategy to enhance production. In our previous research, we conducted an adaptive evolution experiment with Escherichia coli growing under gradually increasing concentrations of ethanol, which gave rise to multiple hypertolerant populations. Based on the genomic mutational data, we demonstrated in this work that adaptive alleles in the EnvZ-OmpR two-component system drive the development of ethanol tolerance in E. coli. Specifically, when a single leucine was substituted for a proline residue within the periplasmic domain using CRISPR, the mutated EnvZ osmosensor caused a significant increase in ethanol tolerance. Through promoter fusion assays, we showed that this particular mutation stabilizes EnvZ in a kinase-dominating state, which reprograms signal transduction involving its cognate OmpR response regulator. Whole-genome proteomics analysis revealed that this altered signaling pathway predominantly maintains outer membrane stability by upregulating global porin levels and attenuating iron metabolism in the tolerant envZ*L116P mutant. Moreover, we demonstrated that the hypertolerant envZ*L116P allele also promotes ethanol productivity in fermentation, providing valuable insights for enhancing industrial ethanol production.

|
Scooped by
?
May 2, 12:32 AM
|
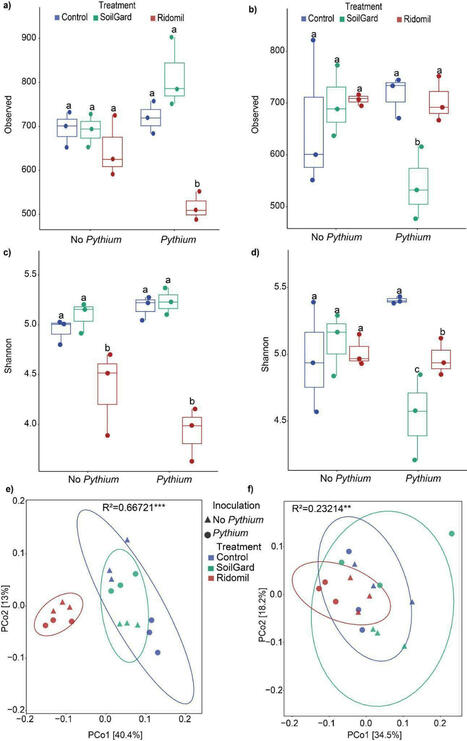
Soil microbial communities play key roles in agroecosystems, particularly in processes like organic matter decomposition and nutrient cycling. However, human activities can negatively impact their community structure and, consequently, soil function. SoilGard and Ridomil are effective methods for controlling carrot cavity spots caused by Pythium spp., but their effects on bacterial taxonomic and metabolic function shifts are not well understood. This study aims to investigate the comparative impact of the chemical fungicide Ridomil and the biological fungicide SoilGard on the bacterial communities in soils cultivated with carrots. Our results showed that both SoilGard and Ridomil significantly impacted soil bacterial diversity, but their effects were distinct and time-dependent. Ridomil had an immediate negative effect on soil bacterial diversity two weeks after treatment, whereas SoilGard was initially less disruptive but showed delayed negative consequences 12 weeks after treatment, particularly when combined with Pythium inoculation. Ridomil treatment led to an increase in Proteobacteria, especially the Pseudomonas population, as confirmed by both MiSeq and qPCR data. In contrast, SoilGard depleted the Mycobacterium population at 12 weeks after treatment. Furthermore, the results of community-level physiological profiling using Biolog Ecoplates showed significant differences in substrate-level diversity between Ridomil and SoilGard-treated samples, indicating a shift in the metabolic activity of bacterial communities. Ridomil-treated samples showed the lowest metabolic activity of bacterial communities, based on the diversity and richness of carbon source utilization, compared to control. Overall, this research highlights the distinct and time-dependent effects of biological and chemical fungicides on soil bacterial communities when applied at recommended doses.

|
Scooped by
?
May 2, 12:21 AM
|
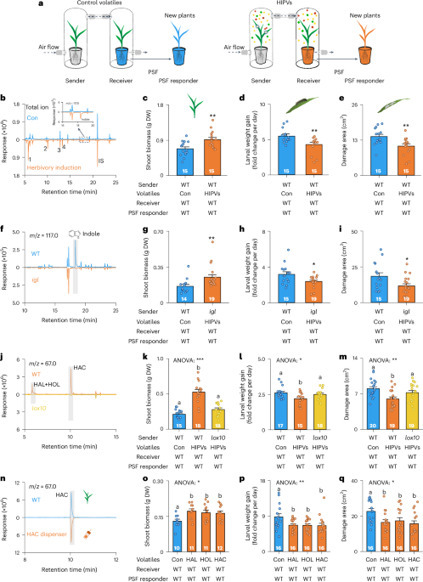
Plants influence each other chemically by releasing leaf volatiles and root exudates, but whether and how these two phenomena interact remains unknown. Here we demonstrate that volatiles that are released by herbivore-attacked leaves trigger plant–soil feedbacks, resulting in increased performance of different plant species. We show that this phenomenon is due to green leaf volatiles that induce jasmonate-dependent systemic defence signalling in receiver plants, which results in the accumulation of beneficial soil bacteria in the rhizosphere. These soil bacteria then increase plant growth and enhance plant defences. In maize, a cysteine-rich receptor-like protein kinase, ZmCRK25, is required for this effect. In four successive year-field experiments, we demonstrate that this phenomenon can suppress leaf herbivore abundance and enhance maize growth and yield. Thus, volatile-mediated plant–plant interactions trigger plant–soil feedbacks that shape plant performance across different plant species through broadly conserved defence signalling mechanisms and changes in soil microbiota. This phenomenon expands the repertoire of biologically relevant plant–plant interactions in space and time and holds promise for the sustainable intensification of agriculture. This study reports that herbivory-induced volatiles can trigger plant–soil feedbacks that boost performance and yields of cereals in the field.

|
Scooped by
?
May 2, 12:08 AM
|
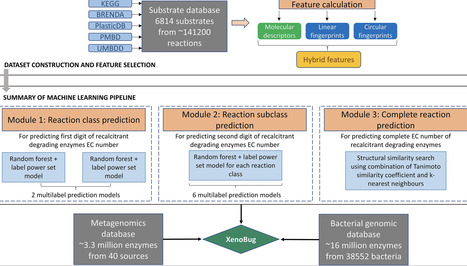
Application of machine learning-based methods to identify novel bacterial enzymes capable of degrading a wide range of xenobiotics offers enormous potential for bioremediation of toxic and carcinogenic recalcitrant xenobiotics such as pesticides, plastics, petroleum, and pharmacological products that adversely impact ecology and health. Using 6814 diverse substrates involved in ∼141 200 biochemical reactions, we have developed ‘XenoBug’, a machine learning-based tool that predicts bacterial enzymes, enzymatic reaction, the species capable of biodegrading xenobiotics, and the metagenomic source of the predicted enzymes. For training, a hybrid feature set was used that comprises 1603 molecular descriptors and linear and circular fingerprints. It also includes enzyme datasets consisting of ∼3.3 million enzyme sequences derived from an environmental metagenome database and ∼16 million enzymes from ∼38 000 bacterial genomes. For different reaction classes, XenoBug shows very high binary accuracies (>0.75) and F1 scores (>0.62). XenoBug is also validated on a set of diverse classes of xenobiotics such as pesticides, environmental pollutants, pharmacological products, and hydrocarbons known to be degraded by the bacterial enzymes. XenoBug predicted known as well as previously unreported metabolic enzymes for the degradation of molecules in the validation set, thus showing its broad utility to predict the metabolism of any input xenobiotic molecules. XenoBug is available on: https://metabiosys.iiserb.ac.in/xenobug.

|
Scooped by
?
May 1, 11:22 PM
|
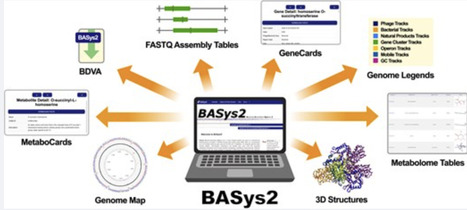
Originally released in 2005, BASys (Bacterial Annotation System) was one of the first web servers to support online bacterial genome annotation and interactive genomic display. Over the past 20 years, web technologies and annotation algorithms have advanced considerably. To keep current with these advances and changing needs of microbial genomics, we have developed BASys2 (Bacterial Annotation System 2.0). BASys2 represents a significant upgrade to BASys, offering much more rapid (up to 8000× faster) and far more complete (2× as many data fields) genome annotation with significantly improved genome visualization capabilities. More specifically, BASys2 reduces annotation time from 24 h to as little as 10 s through a fast genome-matching and a novel annotation transfer strategy. Accepting either FASTA or FASTQ files, BASys2 is able to generate up to 62 annotation fields per gene/protein, leveraging over 30 bioinformatics tools and 10 different databases. Among the more unique features of BASys2 is its extensive support for whole metabolome annotation and complete structural proteome generation. BASys2’s new interactive genome viewer allows rapid, dynamic visualization of complete bacterial genome maps with options to display/hide multiple concentric annotation tracks, show/remove color-coded legends, manipulate the genome map, and select/view individual gene and metabolite annotations. Available as a web server, a desktop viewer application, and a locally installable Docker image, BASys2 allows researchers to achieve unprecedented annotation depth and to easily upload, download, and display these rich genome/metabolome annotations. The BASys2 web server is freely accessible at https://basys2.ca.

|
Scooped by
?
May 1, 10:53 PM
|
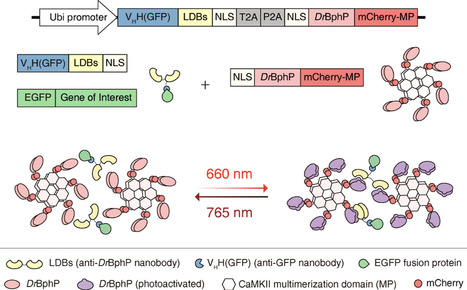
Red light, characterized by superior tissue penetration and minimal phototoxicity, represents an ideal wavelength for optogenetic applications. However, the existing tools for reversible protein inhibition by red light remain limited. Here, we introduce R-LARIAT (red light-activated reversible inhibition by assembled trap), a novel optogenetic system enabling precise spatiotemporal control of protein function via 660 nm red-light-induced protein clustering. Our system harnesses the rapid and reversible binding of engineered light-dependent binders (LDBs) to the bacterial phytochrome DrBphP, which utilizes the endogenous mammalian biliverdin chromophore for red light absorption. By fusing LDBs with single-domain antibodies targeting epitope-tagged proteins (e.g., GFP), R-LARIAT enables the rapid sequestration of diverse proteins into light-responsive clusters. This approach demonstrates high light sensitivity, clustering efficiency, and sustained stability. As a proof of concept, R-LARIAT-mediated sequestration of tubulin inhibits cell cycle progression in HeLa cells. This system expands the optogenetic toolbox for studying dynamic biological processes with high spatial and temporal resolution and holds the potential for applications in living tissues.

|
Scooped by
?
May 1, 10:10 PM
|
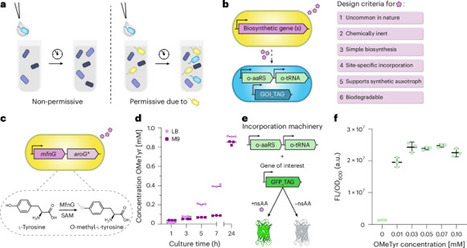
Microorganisms can be genetically engineered for intrinsic biological containment based on synthetic chemical provision. However, reliance on an exogenous chemical limits the contexts where a contained microorganism could survive. Here we design an orthogonal obligate commensalism in Escherichia coli that autonomously creates environments permissive for survival of a partner microbe. We engineer one E. coli strain (the producer) to biosynthesize a non-standard amino acid (nsAA) from simple carbon sources through heterologous expression. We engineer a second E. coli strain (the utilizer) to rely on the same nsAA for growth as a synthetic auxotroph, with a 14-day escape rate of 2.8 × 10−9 escapees per colony-forming unit. Co-culture experiments show utilizer dependence on the producer, with no escape detected during co-inoculation of ~107 colony-forming units of utilizer and a non-producer E. coli strain. Dependence is maintained within a simplified synthetic maize root-associated community. This work provides ecological insights and presents a potential biocontainment strategy independent of an exogenous chemical. A genetically engineered microbial symbiosis based on synthetic auxotrophy is created, with potential implications as a biocontainment strategy.
|
 Your new post is loading...
Your new post is loading...



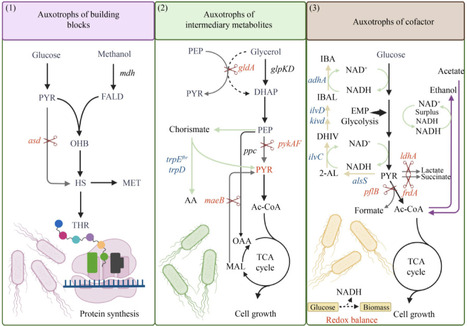

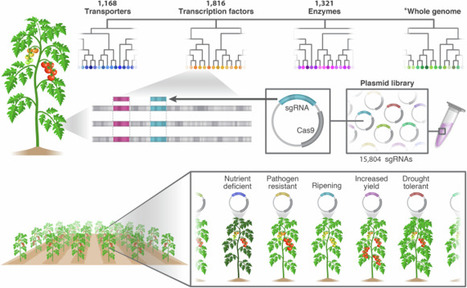
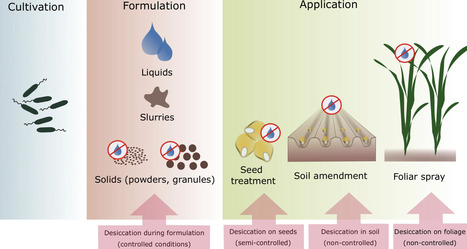








1str, growth coupled metabolite production, auxotrophy