 Your new post is loading...

|
Scooped by
?
Today, 4:59 PM
|
Pseudomonas aeruginosa is a highly adaptable Gram-negative pathogen known for its remarkable ability of forming biofilms. Understanding the environmental cues and regulatory mechanisms that drive biofilm formation is essential for developing effective control strategies. In this study, we screened 57 clinical and environmental P. aeruginosa isolates and discovered that a universal environmental cue, temperature downshift from host-associated 37°C to room temperature (21°C), significantly promotes biofilm formation in 63% of strains. Using the ATCC 27853 strain as a model, we demonstrate that this enhancement results from increased production of the Psl exopolysaccharides at lower temperature. LC-MS/MS analysis revealed elevated levels of the secondary messenger c-di-GMP, a key regulator of the motile-to-sessile transition, at room temperature. Through screening a mutant library targeting 18 c-di-GMP metabolic enzymes, we identified the diguanylate cyclase SiaD within the SiaABCD signaling and functional module as a principal driver of c-di-GMP elevation and biofilm promotion. Further investigation showed that the entire SiaABCD module, especially the signal-sensing domain of SiaA, mediates the temperature-dependent response. Integrating lipidomics with genetics and physiological assays, we show that a temperature downshift triggers rapid membrane perturbations that activate the SiaABCD signaling module, thereby increasing Psl production to strengthen surface adhesion and drive robust biofilm formation. These findings establish temperature downshift as a previously unrecognized physiological cue that promotes biofilm formation in P. aeruginosa, and define an adaptive regulatory pathway linking specific environmental stresses of membrane perturbation to dedicated c-di-GMP signaling module, paving the way for new strategies to disrupt biofilm-associated infections and transmission.

|
Scooped by
?
Today, 4:46 PM
|
The development of robust, food-grade microbial chassis with tailored metabolic functions is critical for advancing synthetic biology applications in health and nutrition. Here, we report a dual genome engineering strategy that integrates CRISPR-Cas9-mediated knock-in with Cre/loxP-driven genome reduction to streamline the genome of Lactococcus lactis NZ9000 and enable stable expression of a high-activity uricase variant. The resulting strain, NZ9000::UAT-ΔD6, demonstrated enhanced enzymatic performance in vitro, achieving 2.34 U/mL activity and complete degradation of ∼500 μM urate within 20 h. Beyond improved catalytic output, this dual-system approach established a genetically stable and biosafe probiotic chassis with moderate colonization capacity in the murine gut. The integration of CRISPR-Cas and Cre/loxP techniques in this work is intended to enhance the expression of heterologous genes in the chassis strain, while providing a versatile platform for the rational design of food-grade probiotics and offering a general strategy for constructing living biotherapeutic agents with targeted metabolic activities.

|
Scooped by
?
Today, 2:53 PM
|
The development of antimicrobial treatments as alternatives to antibiotics to combat antimicrobial-resistant (AMR) bacteria is a global priority. Antimicrobial peptides and proteins such as Host Defense Peptides (HDPs) and endolysins are one of the alternatives that are being explored. HDPs are small, cationic, and amphiphilic antimicrobial peptides derived from the innate immune system exhibiting a broad-spectrum antimicrobial activity. On the other hand, endolysins are enzymes produced by bacteriophages to hydrolyze the bacterial peptidoglycan layer, offering more specific antimicrobial activity than HDPs. While short peptides can be chemically synthesized, this approach presents several limitations, and recombinant production is also being used. Escherichia coli is the most used bacterial expression system for protein production. Alternative systems based on Generally Recognized as Safe (GRAS) microorganisms such as Lactic Acid Bacteria (LAB) have also been employed. However, so far, no comparative studies have evaluated the production and activity of antimicrobial proteins expressed in E. coli versus LAB and this study aims to address that gap. To evaluate potential differences in the production of antimicrobial proteins using E. coli and two LAB (Lactococcus lactis and Lactiplantibacillus plantarum) hosts, various proteins were evaluated. These included two HDPs fused to a GFP, two multidomain HDP-based proteins and one endolysin. The results revealed a clear influence of the expression system on the quality of HDP-based protein, including both GFP fusions and multidomain constructs. Protein yield was higher in E. coli and all HDP-based proteins exhibited higher antimicrobial activity when expressed in E. coli compared to L. lactis and L. plantarum. In contrast, endolysin activity was comparable when produced in E. coli and L. lactis. These results demonstrate that the choice of bacterial expression host significantly affects not only the yield but, more importantly, the antimicrobial activity of HDP-based proteins. For these proteins, the antimicrobial activity was consistently higher when produced in E. coli. In contrast, endolysins exhibited similar characteristics regardless of whether they were expressed in E. coli or in L. lactis.

|
Scooped by
?
Today, 10:22 AM
|
Extreme environments impose strong mutation and selection pressures that drive distinctive, yet understudied, genomic adaptations in extremophiles. In this study, we identify 15 bacterium–archaeon pairs that exhibit highly similar k-mer-based genomic signatures despite maximal taxonomic divergence, suggesting that shared environmental conditions can produce convergent, genome-wide sequence patterns that transcend evolutionary distance. To uncover these patterns, we developed a computational pipeline to select a composite genome proxy assembled from noncontiguous subsequences of the genome. Using supervised machine learning on a curated dataset of 693 extremophile microbial genomes, we found that 6-mers and 100 kbp genome proxy lengths provide the best balance between classification accuracy and computational efficiency. Our results provide conclusive evidence of the pervasive nature of k-mer-based patterns across the genome, and uncover the presence of taxonomic and environmental components that persist across all regions of the genome. The 15 bacterium–archaeon pairs identified by our method as having similar genomic signatures were validated through multiple independent analyses, including 3-mer frequency profile comparisons, phenotypic trait similarity, and geographic co-occurrence data. These complementary validations confirmed that extreme environmental pressures can override traditionally recognized taxonomic components at the whole-genome level. Together, these findings reveal that adaptation to extreme conditions can carry robust, taxonomic domain-spanning imprints on microbial genomes, offering new insight into the relationship between environmental impacts and genome sequence composition convergence.

|
Scooped by
?
Today, 10:13 AM
|
Systematically designing regulatory elements for precise gene expression control remains a central challenge in genomics and synthetic biology. Here we introduce DNA-Diffusion, a generative artificial intelligence framework that uses machine learning trained on DNA accessibility data from diverse cell lines to design compact regulatory elements with cell-type-specific activity. We show that DNA-Diffusion generates 200-base-pair synthetic elements that recapitulate endogenous transcription factor binding grammar while exhibiting enhanced cell-type specificity. We validated these elements using a 5,850-element STARR-seq library across three cell lines. Moreover, we demonstrated successful endogenous gene modulation using EXTRA-seq, reactivating AXIN2, a leukemia-protective gene, in its native genomic context. Our approach outperforms existing computational methods in balancing functional activity with cell-type specificity while maintaining sequence diversity. This work establishes DNA-Diffusion as a powerful tool for engineering compact, highly specific regulatory elements crucial for advancing gene therapies and understanding gene regulation. The authors present DNA-Diffusion, a generative AI framework that designs synthetic regulatory elements with tunable cell-type specificity. Experimental validation demonstrates their ability to reactivate AXIN2 expression, a leukemia-protective gene, in its native genomic context.

|
Scooped by
?
Today, 10:03 AM
|
The 2026 Nucleic Acids Research database issue has 182 papers from across biology and neighboring fields. Eighty-four of these papers describe new databases, while 86 are updates on databases that have previously appeared here. Twelve more papers cover databases most recently published elsewhere. New nucleic acid databases include NapRNAdb for noncapped RNA and GlycoRNAdb. Protein structure is covered by updates from wwPDB members and the AlphaFold Database; SMART, PROSITE, and eggNOG cover domains and families. The Open Enzyme Database and QSproteome are new community-orientated initiatives. JoGo covers hierarchically named and contextualized human haplotypes in the issue’s first Breakthrough paper; So3D provides genuinely 3D spatial transcriptomics in the other. Foundational databases Genenames.org and Gene Ontology also provide updates. The Database Issue is freely available on the Nucleic Acids Research website (https://academic.oup.com/nar). At the NAR online Molecular Biology Database Collection (http://www.oxfordjournals.org/nar/database/c/), over the past year, 899 entries were reviewed, 96 new resources added, and 319 discontinued URLs removed, bringing the total number of databases to 2173.

|
Scooped by
?
Today, 1:55 AM
|
Plant cell walls harbor vast carbohydrate reserves, yet how pathogens unlock them remains unclear. We show that the citrus canker pathogen Xanthomonas citri subsp. citri (Xcc) mobilizes cell wall sugars by hijacking a fruit-ripening program through the type III effector PthA4, which activates the ripening coordinator CsLOB1. CsLOB1 induces approximately 100 genes, many encoding enzymes involved in cell wall breakdown. In the nonfruiting species Nicotiana benthamiana, CsLOB1 likewise promotes Xanthomonas growth, showing that its activity is not strictly dependent on a ripening program. Transcriptomics and reporter assays revealed PthA4-dependent activation of the Xcc xylan CUT system, triggered by host-derived xylose and including a type II–secreted xylanase. Thus, PthA4-driven cell wall remodeling activates bacterial xylan use, establishing a TIII–TII effector feedforward loop that fuels Xcc proliferation.

|
Scooped by
?
Today, 1:35 AM
|
Enzymes are biological catalysts that speed up chemical reactions in an eco-friendly way. Precise enzyme design is hindered by vast sequence space and intricate sequence–structure–function interdependencies. To address these challenges, we developed EvoZymePro-Cat (EZPro-Cat), a deep learning platform for enzyme mutant screening. Conventional methods for predicting absolute mutant activities suffer from systematic errors and limited generalizability. Our pairwise comparison framework directly models relative activity superiority between variants, eliminating dependence on absolute value predictions. The framework integrates full sequence and local structure semantics of protein and ligand information using bilinear attention mechanisms. Protein sequences are encoded using the ESM1b transformer model. Ligands are represented through MolT5 embeddings and MACCS molecular fingerprints. The adaptability of protein residues to their microenvironments is captured by integrating structural features and site-specific evolutionary characteristics. Bilinear attention mechanisms capture long-range intermolecular interactions during catalysis by bidirectional projection and weighted fusion of protein–ligand features. Compared to existing methods, our model exhibits superior performance in identifying improved enzyme mutants through comparative prediction of mutation effects on activity, such as Km and kcat. For deep mutation scanning data sets, a few-shot learning strategy combined with the EZPro-Cat framework boosts prediction precision (AUC 0.908). By using integrated multimodal representations, EZPro-Cat offers a mechanistic and practical solution for functional profiling of intraprotein variants, driving paradigm shifts in highly efficient enzyme discovery and directed evolution.

|
Scooped by
?
Today, 1:25 AM
|
Arctic permafrosts are rapidly thawing in response to climate change, stimulating microbial activity and release of additional greenhouse gases, such as methane. Recently, catechin amendment of thawed permafrost soil microcosms demonstrated >80% decrease in methane production over 35 days compared to unamended controls, with metagenome, metatranscriptome and metabolome analyses revealing a shift in prokaryotic carbon metabolism from a syntrophic network feeding methanogens to one dominated by catechin fermentation. Here we leverage these same data to investigate potential virus impacts on this microbial community metabolism shift. In total, 900 DNA virus operational taxonomic units (vOTUs) were identified as actively lytic based on transcribed structural and lysis genes. Of these, 41% were predicted to infect at least one of 56 transcriptionally active prokaryote genera representing 13 phyla. The most active vOTUs were predicted to infect key catechin-degrading genera including Clostridium and undescribed Bacillota genus JAGFXR01. Temporally, viral communities responded later than prokaryotes to catechin amendment, reflecting a virus production lag to targeting key microbial responders. An induced prophage represented by a vOTU predicted to infect JAGFXR01 dominated the catechin-amended viral community response. It reached abundances 20-156 times higher than its host, suggesting intense viral lysis that could release degraded catechin intermediates. Indeed, catechin intermediate gene expression in non-catechin-degraders were elevated in catechin-amended samples, suggesting JAGFXR01 lysis products were taken up by non-catechin-degraders as part of community carbon metabolism rewiring. Together, these results potentially place viruses at the heart of carbon rewiring that modulates ecosystem outputs of climate-critical thawing permafrosts.

|
Scooped by
?
Today, 1:18 AM
|
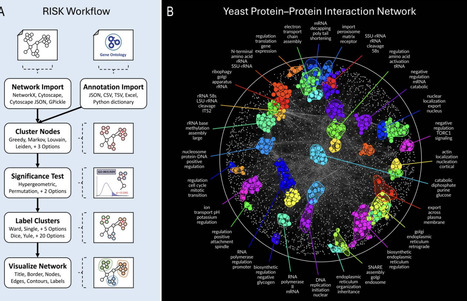
Analyzing biological networks demands scalable annotation tools, yet existing methods fall short in clustering power, statistical flexibility, and broad data compatibility. We introduce RISK (Regional Inference of Significant Kinships), a next-generation tool that overcomes these challenges by integrating community detection algorithms, rigorous overrepresentation analysis, and a modular architecture that supports diverse network types. RISK identifies biologically coherent relationships within networks and generates publication-ready visualizations, as demonstrated by its ability to resolve compact functional modules in Saccharomyces cerevisiae protein–protein interaction and genetic interaction networks. Its application to a high-energy physics citation network reveals structured relationships among research subfields, highlighting its versatility beyond biological systems. As biological and interdisciplinary networks increase in size and complexity, RISK’s scalability and adaptability make it a powerful solution for modern network analysis.

|
Scooped by
?
Today, 1:13 AM
|
Marine environments are frequently oligotrophic, characterized by low amount of bioassimilable nitrogen sources. At the global scale, the microbial fixation of N₂, or diazotrophy, represents the primary source of fixed nitrogen in pelagic marine ecosystems, playing a key role in supporting primary production and driving the export of organic matter to the deep ocean. However, given the high energetic cost of N₂ fixation, the active release of fixed nitrogen by diazotrophs appears counterintuitive, suggesting the existence of alternative passive release pathways that remain understudied to date. Here, we show that the marine non-cyanobacterial diazotroph Vibrio diazotrophicus is endowed with a prophage belonging to the Myoviridae family, whose expression is induced under anoxic and biofilm-forming conditions. We demonstrate that this prophage can spontaneously excise from the genome of its host and that it forms intact and infective phage particles. Moreover, phage-mediated host cell lysis leads to increased biofilm production compared with a prophage-free derivative mutant and to increased release of dissolved organic carbon and ammonium. Altogether, the results suggest that viruses may play a previously unrecognized role in oceanic ecosystem dynamics by structuring microhabitats suitable for diazotrophy and by contributing to the recycling of (in)organic matter.

|
Scooped by
?
Today, 12:47 AM
|
Pooled CRISPR screens using single-cell RNA sequencing (scRNA-seq) have emerged as powerful tools to uncover gene function, map regulatory networks, and identify genetic interactions. However, the inherent sparsity of bacterial scRNA-seq data has posed a major challenge toward applying these approaches to bacteria. Here, we present mapSPLiT, a pooled bacterial CRISPR activation and interference (CRISPRa/i) screening platform that enables large-scale experiments that simultaneously link hundreds of perturbations to their corresponding single-cell transcriptomes. By targeting 52 known or putative transcription factors with 118 perturbations in a pooled experiment, we expanded the E. coli regulatory network map, determined the function of putative regulators, and identified emergent global phenotypes. By targeting combinations of transcription factors simultaneously, we uncovered genetic interactions and regulatory logic between them. Mapping regulatory networks for carbon utilization in P. putida revealed control points that could expand metabolic flexibility and improve biomanufacturing. Together, these results establish mapSPLiT as a generalizable platform for single‑cell functional genomics in bacteria.

|
Scooped by
?
Today, 12:30 AM
|
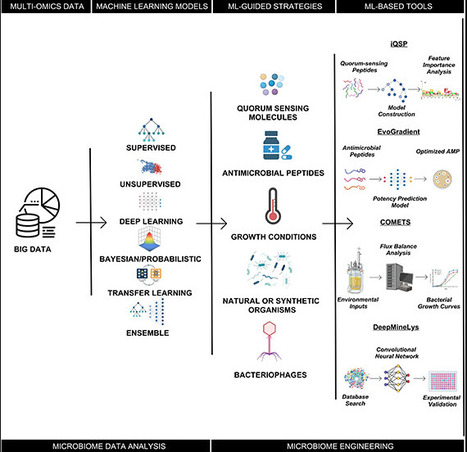
Microbiomes, complex communities of microorganisms and their genetic material, hold immense potential for addressing global challenges in diverse sectors, including healthcare, agriculture, and bioproduction. Engineering these intricate ecosystems, however, necessitates a comprehensive understanding of the complex web of microbial interactions. The emergence of machine learning (ML) has revolutionized microbiome research, offering powerful tools to analyze massive data sets, uncover hidden patterns, and predict microbial behavior. ML algorithms have demonstrated remarkable success in identifying and characterizing microbial communities, predicting interactions between organisms and optimizing the design of microbial communities for specific functions. This Perspective examines the transformative applications of ML in the context of microbiome engineering, encompassing both microbiome data analysis and the targeted manipulation of microbial communities. These techniques employ a variety of strategies, including the manipulation of quorum sensing molecules, antimicrobial peptides, growth conditions, the introduction of probiotics, and the utilization of bacteriophages. By integrating ML with experimental approaches, researchers are pushing the boundaries of microbiome engineering, paving the way for novel applications in diverse fields. However, it is important to acknowledge the challenges that ML algorithms face, such as the limited availability of high-quality, large-scale data sets, the inherent complexity of biological systems, and the need for improved integration of experimental and computational methods. This perspective further discusses the future perspectives of the field, highlighting expected developments in data generation, algorithm development, and interdisciplinary collaboration. These advancements hold the key to unlocking the full potential of microbial communities for addressing pressing global challenges.
|

|
Scooped by
?
Today, 4:52 PM
|
Lactoferrin (LF) plays a positive role in attenuating aging. In this study, LF obtained using different processing methods (freeze-dried: F and spray-dried: S) and its gastrointestinal digesta (XF and XS) were supplemented in d-gal-induced mice to explore their antiaging effects. The results showed that LF and its digesta (LFs) effectively ameliorated cognitive decline. Mechanistically, LFs prevented neuronal and synaptic injury by restoring redox balance, inhibiting the activation of microglia and astrocytes, and activating the cAMP-response element binding protein (CREB)/brain-derived neurotrophic factor (BDNF) pathway. Additionally, LFs increased the tight junction proteins and mucin-2, regulated the gut microbiota, particularly enriching bacteria in Firmicutes and restoring the Firmicutes/Bacteroidota ratio to maintain intestinal homeostasis. Meanwhile, LFs altered phospholipids (PLs) and other metabolites involved in glycerophospholipid metabolism such as arachidonic acid. Correlation analysis showed a significant association among metabolites, microbiota, and behaviors. These results indicated that LF and especially its digesta exert antiaging effects through multitarget pathways involving neuronal protection, neuroinflammation suppression, and microbiota–gut–brain axis regulation.

|
Scooped by
?
Today, 4:40 PM
|
Rubisco catalyzes the CO2 fixation step in the dark reactions of photosynthesis. Transgenic expression of better-performing Rubisco orthologs in plants or discovery of improved mutants of Rubisco via protein engineering could theoretically accelerate plant growth and improve crop yields. However, efforts to heterologously express or engineer Rubisco are frequently stymied by the chaperone-dependent folding and assembly of the Rubisco holoenzyme, a process that can be disrupted by changes to Rubisco’s sequence. Elucidation of the effects that alterations to Rubisco’s sequence impose upon its biogenesis is hampered by reliance upon low-throughput methods for verification of Rubisco assembly. Here, we report the engineering of a genetically encoded biosensor to sense the assembly of Form I Rubiscos in E. coli. We show that the biosensor can detect the RbcS-dependent assembly of cyanobacterial Rubisco orthologs, the formation of chaperone-stabilized RbcL oligomeric assembly intermediates, and differences in assembly caused by mutations to the RbcL sequence. Additionally, we perform a large-scale examination of the relative assembly levels of a ∼7500-member Halothiobacillus neapolitanus RbcL mutant library by adapting the biosensor for use with phage-assisted noncontinuous selection. Our experiment predicts that the majority (>90%) of examined RbcL mutations exert a negative effect on assembly, lending support to the hypothesis that Rubisco biogenesis constrains both its natural evolution and improvement by protein engineering.

|
Scooped by
?
Today, 11:13 AM
|
Recombinant adeno-associated viral (rAAV) vectors are a leading platform for in vivo gene therapy, valued for their excellent safety, broad serotype diversity, and scalable production. Targeted delivery through capsid display of ligands holds great promise, yet current retargeting strategies often rely on extensive capsid re-engineering and restrict the use of ligands incompatible with intracellular expression systems. Here, we present a modular AAV retargeting platform that, for the first time, employs the SpyTag/SpyCatcher system via genetic integration into the AAV2 capsid. SpyTag is a small peptide that forms a covalent, irreversible bond with its protein partner, SpyCatcher, allowing site-specific ligand coupling under physiological conditions. Inserting SpyTag into surface-exposed capsid sites enabled postassembly functionalization of AAVs with SpyCatcher-fused targeting proteins. As proof of concept, we used SpyCatcher fusions with designed ankyrin repeat proteins (DARPins) specific for EGFR, EpCAM, and HER2. This conferred highly specific transduction of corresponding cancer cell lines with minimal off-target activity. Therapeutic potential was demonstrated by delivering a suicide gene, inducing selective cancer cell killing upon prodrug administration. This “one-fits-all” platform allows rapid and flexible retargeting without significantly altering the underlying vectors genome or production process. It supports the incorporation of large or complex ligands not amenable to genetic fusion and facilitates high-throughput preclinical evaluation strategies. By uniting capsid engineering with modular ligand display, our approach provides a scalable and versatile framework for precision gene delivery, broadening the applicability of rAAV in both therapeutic and discovery settings.

|
Scooped by
?
Today, 10:17 AM
|
Bilirubin biosynthesis has long been constrained by low yields and poorly understood bottlenecks. Here, we report a fully in vitro pathway that converts heme to bilirubin with the titer of 1.7 g/L and 95.8%. Systematically, enzyme screening and mechanistic analysis reveal the hidden challenge: Fe²⁺-induced oxidative degradation of intermediates. We show that Fe²⁺ coordinates with deprotonated intermediates to trigger oxidative ring-opening degradation via O₂-mediated radical mechanism, as supported by DFT calculations indicating reduced HOMO-LUMO gap in Fe²⁺-ligand complexes. The degradation is mitigated through competitive iron chelation and protonation state modulation, improving yield to 80.1%. Furthermore, we have resolved heme-CO complexes blocking O₂ activation at heme oxygenase by introducing a carbon monoxide dehydrogenase to remove CO and restore enzyme activity. Coupled with NADPH-recycling via formate dehydrogenase, these interventions enable efficient, scalable bilirubin synthesis with a 20-fold improvement. Our work shows controlling inhibitory byproducts is critical for stabilizing heme-related pathways and as a generalizable framework for synthetic biology. The large-scale production of bilirubin has long been a challenge. Here, the authors engineered an in vitro enzymatic cascade through systems-level design, enabling gram-scale synthesis of bilirubin.

|
Scooped by
?
Today, 10:09 AM
|
The development of sustainable nanobiocatalysts is a focal challenge in green chemistry, requiring robust and eco-friendly production methods. This study introduces a single-source strategy that addresses this challenge. By utilizing the fungus Aspergillus niger as a single biological factory to simultaneously produce a lipase enzyme and biosynthesize the iron oxide nanoparticles (IONPs) that serve as lipase matrix support. This integrated approach ensures a high degree of compatibility between the enzyme and its nanoparticle support, which was confirmed during the immobilization step by an 81.73% yield and a remarkable 97.4% activity retention. The resulting nanobiocatalyst demonstrated a distinct operational stability, broad pH tolerance, and maintained over 80% of its activity after eight consecutive reuse cycles. In practical applications, the catalyst showed powerful bioremediation capabilities, achieving near-complete (> 95%) removal of industrial dyes and effective oil-stain removal from fabric. Our findings could establish that using a single biological source for both the enzyme and its immobilization matrix offers a new benchmark for future enzyme immobilization technologies.

|
Scooped by
?
Today, 9:59 AM
|
Methanogenic archaea emit ~1 Gt of methane annually, impacting global carbon cycling and climate. Central to their energy metabolism is a membrane-bound, sodium-translocating methyltransferase complex: the N⁵-tetrahydromethanopterin:CoM-S-methyltransferase (Mtr). It couples methyl transfer between two methanogen-specific cofactors with sodium ion transport across the membrane, forming the only energy-conserving step in hydrogenotrophic methanogenesis. Here, we present a 2.1 Å single-particle cryo-EM structure of the Mtr complex from Methanosarcina mazei. The structure reveals the organization of all catalytic subunits, embedded archaeal lipids and the sodium-binding site. Most strikingly, we discover MtrI, a previously unannotated small open-reading frame encoded protein ( < 100 aa) found within the order of Methanosarcinales that binds both the top of the sodium-channel and cytosolic domain of MtrA via its cobamide cofactor in response to oxygen exposure. This interaction likely prevents sodium leakage and stabilizes the complex under oxidative conditions, revealing an unexpected regulatory mechanism in methanogen energy conservation. Here, the authors present the cryoEM structure of the sodium-translocating methyltransferase (Mtr) complex from Methanosarcina mazei. Along with providing catalytic insights, they identify MtrI, an unannotated small protein, bound to the Mtr complex in a redox-dependent manner.

|
Scooped by
?
Today, 1:52 AM
|
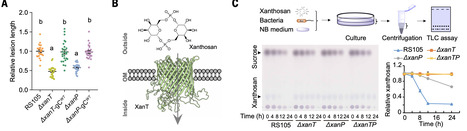
Xanthomonas spp. cause serious diseases in more than 400 plant species. The conserved AvrBs2 family effectors are among the most important virulence factors in xanthomonads, but how AvrBs2 promotes infection remains elusive. We found that AvrBs2 is a glycerophosphodiesterase-derived synthetase that catalyzes uridine 5′-diphosphate-α-d-galactose into a sugar phosphodiester, bis-(1,6)-cyclic dimeric α-d-galactose-phosphate, which is referred to as xanthosan. Xanthosan is synthesized by AvrBs2 in host cells and released into apoplastic spaces. Xanthomonas bacteria uptake xanthosan through the XanT transporter and hydrolyze it through the XanP phosphodiesterase for nutrition. AvrBs2, XanT, and XanP form a xanthosan “generation-uptake-utilization” system to provide a dedicated nutritional strategy to feed xanthomonads. Furthermore, elucidation of the AvrBs2-XanT-XanP virulence mechanism inspired us to develop an “anti-nutrition” strategy that should be applicable to control a wide variety of Xanthomonas diseases.

|
Scooped by
?
Today, 1:29 AM
|
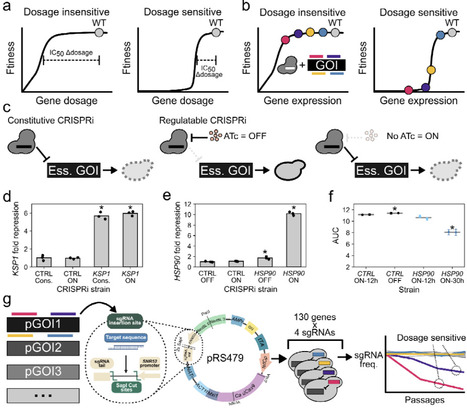
The rising rate of drug-resistant fungal infections and the emergence of fungal pathogens with intrinsic resistance phenotypes are a growing concern. The close evolutionary distance between mammals and fungi complicates the design of new antifungals and increases the chances of toxic off-target effects. As such, antifungal drug development usually focuses on fungal-specific proteins when considering potential new targets. Ideal drug targets should mediate essential cell processes and be highly sensitive to inhibition. Targeted gene repression can serve as a model for drug-mediated inhibition and for determining the dosage-sensitivity profile of genes of interest. In the fungal pathogen Candida albicans, classical approaches for gene repression can be labor-intensive and limited to one genetic background due to low throughput. Here, we adapt pooled CRISPRi in C. albicans for the first time and exploit this technique for large-scale functional genomic analysis. Through pooled CRISPRi screening, we test the repression sensitivity of over a hundred essential genes conserved in fungi but absent in humans in a single flask, and successfully identify highly dosage-sensitive genes across multiple cell components and pathways. We further extended our analysis to ten diverse environmental conditions, allowing us to measure how the environment influences dosage-sensitivity profiles. Finally, we extend our experiments to two clinical drug-resistant C. albicans strain backgrounds and demonstrate that many of the fitness defects we observed are conserved in resistant clinical isolates. Together, our results highlight a set of genes that are highly dosage-sensitive across different genetic and environmental contexts, making them attractive targets for further investigation. By facilitating rapid, efficient large-scale functional genomics assays across diverse genetic backgrounds, CRISPRi pooled screening will open new frontiers in C. albicans biology.

|
Scooped by
?
Today, 1:22 AM
|
Antibiotic persistence, characterized by a dormant subpopulation of bacterial cells that causes chronic and recurrent infections, remains poorly understood despite being recognized nearly a century ago. Toxin–antitoxin (TA) systems, which include a toxin and an antitoxin, are promising candidates for elucidating persister formation. We present the first theoretical model of persister formation driven by type I TA systems, in which the antitoxin is a small RNA molecule. Our analyses and simulations reveal two steady states—low toxin (normal growth) and high toxin (persistence)—with stochastic switching between them. Bistability requires both positive and negative feedback mediated by inhibition of antitoxin degradation. We derive stability diagrams that map mechanistic properties to system dynamics. The model suggests that while type I TA systems may not produce persisters under normal conditions, they can enter a bistable regime under stress, such as antibiotic exposure or nutrient limitation, leading to increased toxin expression or slower growth. Moreover, transiently slow-growing cells can be stabilized as long-living persisters through bistable TA dynamics. Using a cusp catastrophe surface, we identify distinct roles for two toxin inhibition mechanisms in modulating steady states and hysteresis. These findings provide a mechanistic basis for experimental observations and a framework for future studies.

|
Scooped by
?
Today, 1:15 AM
|
Metabolic network modeling is essential for understanding metabolic shifts occurring in complex physio-pathological processes. Currently, constraint-based modeling frameworks for metabolic networks primarily rely on Python or MATLAB libraries, requiring some coding skills. In contrast, more user-friendly tools lack essential features such as flux sampling or transcriptomic data integration We introduce COBRAxy, a Python-based tool suite integrated into the Galaxy Project. COBRAxy enables constraint-based modeling and sampling techniques, allowing users to compute metabolic flux distributions for multiple biological samples. The tool also enables the integration of medium composition information to refine flux predictions. Additionally, COBRAxy provides a user-friendly interface for visualizing significant flux differences between populations on an enriched metabolic map. This extension provides a comprehensive and accessible framework for advanced metabolic analysis, enabling researchers without extensive programming expertise to explore complex metabolic processes.

|
Scooped by
?
Today, 12:59 AM
|
Invertebrates, as the majority of macroscopic species on the Earth, are important resources for natural products. Chemical investigations of animals can date back to the early 20th century and have led to the discovery of thousands of compounds with diverse biological functions. These natural products can be structurally classified as terpenoids, polyketides, and alkaloids. Additionally, many compounds have been isolated from symbionts, leading to the widespread belief that animals lack the capability for secondary metabolism. Recent biochemical studies challenge this notion, revealing great potential for animal biosynthesis research. Animals possess larger genomes and more complex metabolic pathways, suggesting untapped biosynthetic potential. In contrast to microorganisms, studies on the biosynthesis of natural products in animals remain limited. Characterized genes represent only a small fraction of their vast genomes. The discovery of biosynthetic gene clusters suggests that the methods used to mine the biosynthetic genes of microorganisms may also be applicable to animals. The characterization of 4-vinylanisole in locusts demonstrates that the pathways lacking clear core biosynthesis enzymes still require multidisciplinary experimental approaches. In summary, further biosynthesis studies will expand methodological approaches and accelerate the characterization of remaining natural product pathways.

|
Scooped by
?
Today, 12:41 AM
|
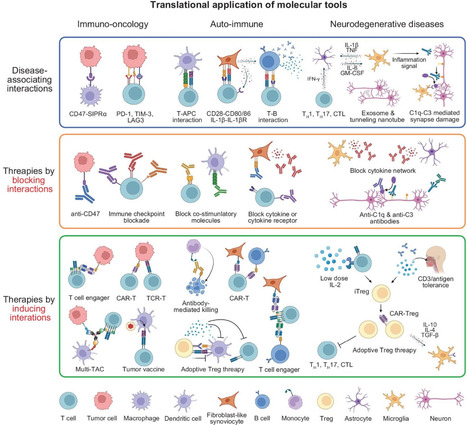
Cell-cell communication (CCC) is fundamental to essential biological processes including growth, differentiation, immune surveillance, and tissue homeostasis, and its dysregulation underlies various diseases such as cancer, autoimmunity, and neurodegeneration. In response to growing interest in decoding complex multicellular interactions, the 380th Shuangqing Forum entitled ‘Chemical, Biological, and Medical Frontiers in Multicellular Complex Systems’ was convened, providing a platform to discuss recent interdisciplinary breakthroughs. This review, emerging from forum discussions, highlights the latest advancements in molecular tools—such as super-resolution imaging, proximity labeling, bioorthogonal chemistry, synthetic receptors, and single-cell spatial omics—that enable unprecedented insights into spatial, molecular, and functional aspects of CCC. Emphasizing their translational potential, we discuss their profound implications for immuno-oncology, regenerative medicine, and autoimmune diseases. We further outline current challenges and opportunities, particularly advocating for a future precision medicine framework centered around targeted modulation of cell-cell interactions.
|
 Your new post is loading...
Your new post is loading...





























gao c.