 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:16 AM
|
Small molecule–regulated protein oligomerization provides a powerful mechanism for manipulating biological processes by controlling protein proximity with high temporal precision. However, such systems only rarely exist in nature and remain a substantial challenge for de novo design. In this work, we describe a computational method for designing protein homooligomers whose assembly is regulated by small-molecule ligands with matching symmetry. We designed protein homotrimers regulated by the Food and Drug Administration (FDA)–approved drug amantadine and further designed amantadine-responsive heterodimers and heterotrimers. Biophysical characterization confirmed their amantadine-dependent assembly, and their crystal structures closely matched the design models. We demonstrated their broad applicability in controlling protein localization, membraneless condensate formation, and gene expression. Our approach opens new avenues for designing small molecule–responsive proteins and expands the chemogenetic toolkit for manipulating complex biological processes.

|
Scooped by
mhryu@live.com
Today, 12:02 AM
|
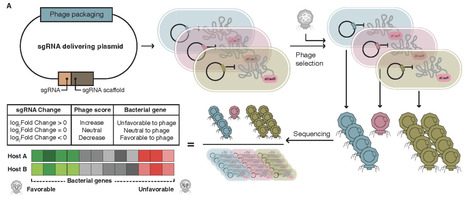
Pseudomonas aeruginosa is a major opportunistic pathogen implicated in a wide range of infections, including chronic respiratory infections, burn wound infections, urinary tract infections, and device-associated infections. Its intrinsic and acquired resistance mechanisms, particularly its capacity for biofilm formation, pose serious challenges to conventional antibiotic therapy. With the continued rise of multidrug-resistant and pan-drug-resistant strains, the need for alternative therapeutic strategies has become increasingly urgent. Phages, viruses that specifically recognize and lyse bacteria, have shown unique advantages in combating antibiotic-resistant infections. This review systematically summarizes recent advances in the application of phage therapy for P. aeruginosa infections, covering in vitro bactericidal activity, biofilm degradation, and synergistic interactions with antibiotics. We further discuss evidence from animal models, including therapeutic efficacy, immunomodulatory effects, and pharmacokinetics. Emphasis is placed on clinical use cases, including different routes of administration, symptom relief, biomarker modulation, pathogen clearance rates, and adverse events. Typical case reports and early-phase clinical trials support the safety and efficacy of phage therapy. Nevertheless, translational barriers persist, such as the need for precise host matching, risks of immune neutralization, and the lack of standardized regulatory frameworks and Good Manufacturing Practice (GMP)-grade production systems. The rapid development of engineered phages and individualized therapeutic approaches offers a feasible path forward. In conclusion, phage therapy holds significant promise for the treatment of drug-resistant P. aeruginosa infections, and future efforts should focus on establishing standardized systems, conducting multicenter clinical studies, and leveraging synthetic biology to accelerate its translation from bench to bedside.

|
Scooped by
mhryu@live.com
January 6, 11:57 PM
|
Acetyl-CoA synthetase (ACS) is a well-characterized enzyme that catalyzes the ATP-dependent ligation of acetate and coenzyme A to produce acetyl-CoA, a central metabolite coordinating energy metabolism, carbon flux distribution, and post-translational protein modification. Recently, ACS has emerged as a metabolic nexus with broad implications for plant–microbe interactions in agriculture. Beyond its canonical role in primary metabolism, ACS governs diverse physiological processes in beneficial plant-associated microorganisms, including rhizosphere colonization, stress adaptation, secondary metabolite biosynthesis, and morphological development—all of which enhance plant growth and resilience. In contrast, in phytopathogens, ACS is closely related to the expression of virulence factors. Thus, ACS exerts a dual influence, shaping both mutualistic and antagonistic microbial lifestyles in planta. This review synthesizes recent advances in the structural and catalytic diversity of ACS, delineates its ecological and functional roles in agriculturally relevant microorganisms, and explores the environmental and host-derived signals that regulates its expression and activity. Particular attention is given to the interplay between ACS-mediated carbon metabolism and protein acetylation, which together modulate microbial physiology and plant-associated behaviors. ACS is thereby positioned as a strategic metabolic hub, providing a framework for future research at the interface of microbial metabolism, environmental adaptation, and plant health.

|
Scooped by
mhryu@live.com
January 6, 11:17 PM
|
Yeasts and yeast-based products are nutrient-rich bioresources with broad applications in technologies for the production of food, feed, medicine, and cosmetics. However, traditional processing often results in non-specific lysis and suboptimal product quality. Yeast extract can be used as a flavor enhancer, nutritional supplement, or fermentation substrate, and the other components of the yeast cell wall and nucleic acids can be processed into bioactive materials, including glucans and nucleotides. These materials offer both nutritional and therapeutic benefits. Precision hydrolysis, leveraging the high specificity of tailored enzymes, has emerged as a superior strategy for maximizing the yield and functional quality of high-value yeast-based products. It provides superior outcomes by improving the quality of yeast-based products. Tailored enzymatic strategies, leveraging mechanistically focused core enzymes, including proteases, β-glucanases, and coupled nucleases-deaminases, have demonstrated superior efficiency, nutritional enhancement, and sensory refinement. This review focuses on the mechanistic properties of yeast processing enzymes, emphasizing their functional classification and applications in precision hydrolysis. It details how such enzymes are optimized for the targeted release and modification of high-value components. Additionally, the review highlights recent strategies for tailored biosynthesis of yeast processing enzymes, including enzyme discovery, heterologous expression systems, and machine-learning-guided optimization. This review aims to support future innovations that will promote the development of sustainable, high-value, and diversified yeast-based bioproducts by optimizing the biosynthesis of processing enzymes, thus lowering the overall cost of precision hydrolysis.

|
Scooped by
mhryu@live.com
January 6, 10:52 PM
|
Microbes process input information into output responses through diverse genetic and metabolic mechanisms, effectively making them physical systems that compute. These computations profoundly shape the environment, from driving key chemical cycles in the soil to influencing the planet’s atmosphere. Yet the complexity of natural microbial computations remains poorly understood, including the symbolic representation of information and the underlying algorithmic principles. Synthetic biology provides tools to implement simple but effective genetic circuits in living cells, enabling human-defined computations. These are typically Boolean gates and circuits for combinatorial input processing, but they also include sequential logic, memory-based systems, analog circuits, and distributed computations in cellular consortia. Twenty-five years after the first synthetic genetic circuits were built, the field is now exploring new approaches to move closer to the computing power of natural microbes. With a focus on bacteria, this review examines both natural and synthetic functions with the aim of bridging the complexity gap between them and argues that understanding and formalizing the ways in which microbes compute may be essential for improving synthetic genetic circuitry.

|
Scooped by
mhryu@live.com
January 6, 4:24 PM
|
The persistence of insecticide residues on food crops poses a significant risk to human health and the environment, necessitating effective residue mitigation strategies. This study evaluates the potential of lactic acid bacteria (LAB) strains, Lactobacillus pentosus and Lactococcus lactis subsp. lactis, for degrading residues of cyantraniliprole and chlorantraniliprole under laboratory conditions, and further assessed their degradation potential for cyantraniliprole under field conditions. Residue levels were quantified using liquid chromatography-tandem mass spectrometry, allowing precise measurement of degradation rates. In minimal broth, both strains reduced cyantraniliprole residues up to 33.07% and chlorantraniliprole residues by up to 23.44% over 12 days, with Lactobacillus pentosus demonstrating a higher degradation efficiency. Nutrient broth significantly enhanced biodegradation efficiency, with both LAB strains removing more than 98% of cyantraniliprole within 4 days and achieving complete degradation by 10th day. Controlled field evaluation demonstrated the practical efficacy of LAB application, where Lactobacillus pentosus and Lactococcus lactis reduced insecticide residues by up to 40.99 and 34.52% respectively, after 8 h of spraying. Overall, these findings highlight the potential of LAB as a natural, eco-friendly solution for mitigating pesticide residues in food commodities, contributing to sustainable agricultural practices.

|
Scooped by
mhryu@live.com
January 6, 4:13 PM
|
Natural products (NPs) are a rich source of therapeutic and agricultural compounds. Unfortunately, many promising metabolites are not expressed under standard laboratory conditions. Deepening our understanding of the regulatory networks governing NP biosynthetic genes is essential for unlocking this hidden chemical diversity. Cluster-situated regulators (CSRs) are transcription factors involved in the regulation of NPs, but their full regulatory range has remained elusive due to limited genome-wide data. Using DNA Affinity Purification Sequencing (DAP-seq), we defined the predicted regulons for 84 CSR homologs across 78 Streptomyces strains. CSRs in this cohort exerted influence across multiple cellular processes, with particularly strong impacts on other transcription factors throughout the genome. Approximately 30% of predicted NP biosynthetic gene clusters (BGCs) contained CSR-regulated genes. In strains encoding multiple CSR homologs, we observed substantial overlap in BGC regulation. Together, these results greatly expand the genomic landscape of CSR activity and provide a foundation for improved bioinformatic strategies to predict and interpret regulatory control of NP biosynthesis.

|
Scooped by
mhryu@live.com
January 6, 4:06 PM
|
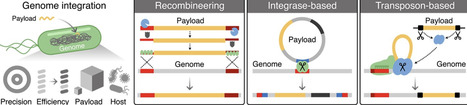
The ability to precisely insert DNA payloads into a genome enables the comprehensive engineering of cellular phenotypes and the creation of new biotechnologies. To achieve such modifications, the most widely used techniques rely on a host cell’s native DNA repair mechanisms like homologous recombination, which hampers their broader use in organisms lacking these capabilities. Here, we explore the current landscape of genome integration systems with a particular focus on those that function in bacteria and are precise, self-contained, and portable, placing minimal requirements on the host cell. Through a historical analysis, we observe long-term use of recombineering technologies, a recent rise in the use of CRISPR-guided systems that consist of associated integrase machinery, and growing efforts to modify non-model organisms. Looking forward, we highlight some of the remaining challenges and how synthetic genomics may offer a way to create bacterial strains optimized for extensive long-term modification. As the field of synthetic biology sets its sights on real-world impact, the effective engineering of genomes will be critical to shaping the robust phenotypes that applications demand.

|
Scooped by
mhryu@live.com
January 6, 4:00 PM
|
Building synthetic versions of biological cells from the bottom up offers an unprecedented opportunity to understand the rules of life and harness cellular capabilities in biotechnology. Whereas substantial progress has been made in recapitulating elementary cell functions, we argue that accelerating the engineering of synthetic cells requires a shift in research practices. The dominant approach—rationally designing and integrating functional modules—becomes restrictive when dealing with the massively complex biochemical pathways associated with life, especially when design principles remain unclear. We advocate moving away from theoretical rational design towards a data-driven model that is centred on library generation. Inspired by a systems chemistry perspective, this strategy prioritizes the systematic creation and distribution of composition–function libraries. To enable this, experimental strategies must integrate high-throughput synthetic cell generation, automation and closed-feedback control of workflows. Broad adoption will also require greater emphasis on quantitative benchmarking, and the de-skilling of techniques, supporting effective laboratory-to-laboratory collaboration. Living cells rely on the choreography of multiple simultaneous functions, without clear boundaries between molecular subsystems. Replicating these capabilities in synthetic cells would represent a major advance in understanding life. This Perspective argues that this challenge requires a shift away from modular design concepts, towards a strategy that integrates the theoretical principles of systems chemistry with data-driven high-throughput experimental methods.

|
Scooped by
mhryu@live.com
January 6, 3:51 PM
|
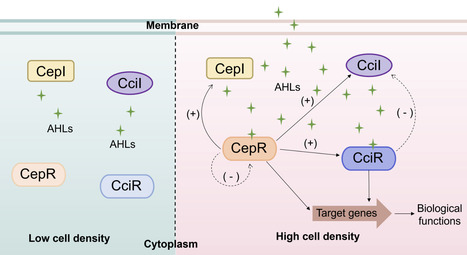
Quorum sensing (QS) is a cell-cell communication mechanism widely employed by bacteria to control group behaviors in a cell density-dependent manner. QS plays a critical role in the regulation of physiological processes in the Burkholderia cepacia complex (Bcc), which consists of at least 28 closely related species. To date, several different QS systems have been identified in the Bcc, including the well-characterized N-acyl-L-homoserine lactone-type QS systems and diffusible signaling factor-type QS systems. Here, we review the research progress on QS in the Bcc, including biosynthesis, biological functions, and regulatory mechanisms. We compare the biosynthetic pathways and regulatory mechanisms of these QS signals, which reveal their specificity and universality. We also review recent antibacterial research, which focuses on targeting these QS signaling systems, and the application prospects of this strategy.

|
Scooped by
mhryu@live.com
January 6, 3:37 PM
|
Ensuring food security while improving vegetable quality remains a major challenge amid population growth and environmental stress. Nano-fertilizers (NFs), with their enhanced bioavailability and regulatory effects on plant metabolism, offer promising solutions to improve nutrient use efficiency, nutritional value, and postharvest quality in vegetables. This paper systematically reviews 106 studies on the effects of NFs under nonbiotic stresses including light, temperature, water, and salinity. Research demonstrates that NFs protect photosynthetic systems; for instance, Se NPs reduce chlorophyll degradation in Chinese cabbage while increasing carotenoid content by 21%. They also enhance antioxidant capacity, reducing malondialdehyde accumulation by over 50%. Nutritionally, NFs promote vitamin synthesis, with zinc oxide NPs increasing vitamin C in tomatoes by 28%, and improve micronutrient levels. Postharvest applications show that NFs inhibit microbial growth and delay ripening, extending shelf life by 30 to 50%. However, significant knowledge gaps remain regarding environmental safety, long-term impacts, and species-specific responses. Future work should prioritize smart-responsive NFs integrated with precision agriculture to reduce fertilizer inputs by 20 to 30% while increasing yields, supporting sustainable vegetable production.

|
Scooped by
mhryu@live.com
January 6, 1:06 AM
|
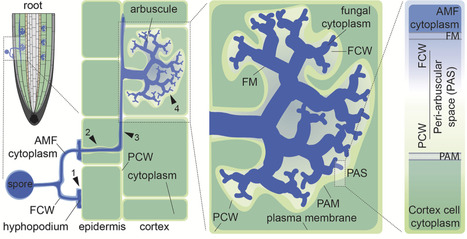
Arbuscular mycorrhizal (AM) associations of plants and Glomeromycotina soil fungi play a crucial role in all terrestrial ecosystems. In this mutually beneficial interaction, obligate biotrophic fungi acquire photosynthetically fixed carbon from the plant, while the mutualistic fungi enhance plant access to soil nutrients. AMF colonize the inner tissues of host roots, where they form specialized symbiotic structures (arbuscules) within fully differentiated cortex cells that are reprogrammed to host the microbe. Given the intimate nature of the interaction, extensive partner communication at the interface of plant and fungal cells is crucial for the development and functioning of AM symbiosis. The peri-arbuscular space, a specialized apoplast compartment surrounding the arbuscules, supports not only nutrient exchange between the symbiotic partners but is also the site of extensive partner crosstalk mediated by cell wall components, receptors, signaling peptides, and extracellular vesicles. Such signaling processes in the apoplast modulate plant immune responses to enable colonization by beneficial fungi, making this compartment a key player for the establishment and maintenance of AM symbiosis. In this review, we discuss recent discoveries related to the role of partner communication in the apoplast, with a focus on peptide and cell wall signaling, as well as extracellular vesicles.

|
Scooped by
mhryu@live.com
January 6, 12:10 AM
|
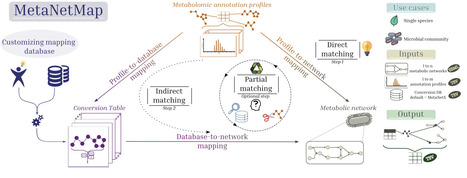
Metabolic networks represent genome-derived information about the biochemical reactions that cells are capable of performing. Mapping omic data onto these networks is important to refine model simulations. However, metabolomic data mapping remains very challenging due to difficulties in identifier reconciliation between annotation profiles and metabolic networks. MetaNetMap is a Python package designed to automatise the process of mapping metabolomic data onto metabolic networks. It includes several layers of identifier matching, the use of customisable databases, and molecular ontology integration to suggest the most matches between experimentally-identified metabolites and molecules defined in the network. We demonstrate its usability and the quality of automated mapping using two datasets.
|

|
Scooped by
mhryu@live.com
Today, 12:11 AM
|
Understanding the extrinsic and intrinsic environmental factors that influence lag phase duration is critical for developing strategies to control the growth of foodborne pathogens. Although the exponential growth rate can be predicted as a straightforward response to the growth environment, lag phase duration is difficult to predict because it depends on not only the current growth conditions but also the previous growth environment and physiological status of the bacterial cells. Therefore, this article aims to provide a comprehensive understanding of the dynamic, adaptable, and evolvable nature of the lag phase. It is based on relevant literature (experimental studies, literature summaries, observational data) on the effect of pre- and postgrowth environments. We discuss the modeling strategies employed to incorporate physiological heterogeneity and dynamic food environments into predictive modeling frameworks. Overall, we summarize the empirical and mechanistic modeling strategies for quantifying lag phase duration.

|
Scooped by
mhryu@live.com
January 6, 11:59 PM
|
New approaches to engineering plant genomes have the potential to improve agriculture. However, transgenes insertion and tissue culture have become bottlenecks to genome-editing technology becoming widely adopted and achieving the promise of targeted editing. Recent developments in particle bombardment and viral vector-mediated delivery can open doors to overcome these limitations.

|
Scooped by
mhryu@live.com
January 6, 11:39 PM
|
Pulmonary drug delivery offers several advantages over systemic administration, including localized targeting of diseased lung tissue, rapid absorption through the extensive alveolar surface area, and minimized systemic side effects owing to reduced off-target distribution. These benefits make it a promising approach for treating a range of respiratory and systemic conditions. However, mucociliary clearance, immune surveillance and enzymatic degradation pose major challenges to drug retention in the lungs. In this Review, we discuss microrobotic delivery systems for tackling the challenges of pulmonary drug delivery. We first outline key considerations for the design of microrobots for pulmonary delivery, including propulsion systems, targeting, controlled drug release, overcoming of biological barriers, delivery routes and clearance, and we then highlight applications for lung-related conditions. In particular, we outline biohybrid platforms based on green algae and inhalable platforms for the treatment of pneumonia and lung metastasis. Finally, we examine the engineering of microrobotic swarms and survey future opportunities and milestones for microrobotic pulmonary delivery. Delivering drugs directly to the lungs enables targeted treatment and long-term drug retention, while minimizing systemic side effects. This Review explores the design of microrobotic platforms engineered for pulmonary drug delivery.

|
Scooped by
mhryu@live.com
January 6, 11:11 PM
|
Spatiotemporal imaging of genomic DNA dynamics in live mammalian cells is essential for elucidating eukaryotic organization and processes relevant to health and disease. CRISPR systems greatly facilitate the development of live cell DNA imaging tools. However, conventional CRISPR imaging tools typically utilize constitutively fluorescent proteins, resulting in high background noise, nonspecific nucleolar signals, and low signal-to-noise ratios. To address this, fluorogenic CRISPR-based imaging tools have been developed. These tools remain non-fluorescent until they bind to the target DNA, thus significantly reducing the background and enhancing the sensitivity. This review summarizes four fluorogenic CRISPR strategies, each utilizing different fluorogenic reporters, including fluorogenic proteins, fluorogenic RNA aptamers, split fluorescent proteins, and molecular beacons. These fluorogenic CRISPR approaches successfully monitored the subnucleus gene loci localization, dynamics, and DNA breaks and repairs. We anticipate that this review can inspire researchers to expand the fluorogenic CRISPR for cellular DNA imaging and diverse bioapplications.

|
Scooped by
mhryu@live.com
January 6, 4:30 PM
|
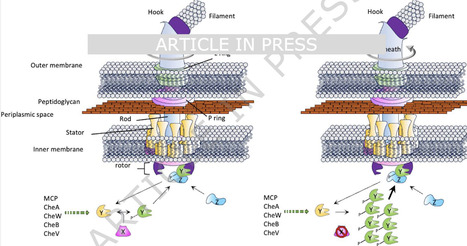
Chemotaxis is an adaptive mechanism that shapes the behavior of motile bacteria in habitats characterized by fluctuating and often conflicting cues environmental (e.g. stay-or-go). Chemotactic responses are orchestrated by phosphorylation of CheY, which triggers rotational switching of the flagella. In Escherichia coli and similar taxa, CheZ is the principal CheY-P phosphatase, whereas in lineages lacking CheZ, members of the structurally distinct CheC-FliY-CheX family fulfill this role. Intriguingly, some bacteria code for CheX and CheZ, presenting a conundrum regarding their function, and the role of CheX in CheZ-containing organisms is unknown. We imposed a sustained motility constraint under conditions of looming nutrient depletion in Vibrio vulnificus, which possesses both CheX and CheZ, using the c-di-GMP effector PlzD that robustly curtails swimming motility. Our analyses revealed that the activity of CheX, but not CheZ, could be attenuated to mitigate the imposed constraint, assigning CheX a pivotal function in fine-tuning foraging behavior during a “stay-or-go” decision. V. vulnificus CheX maintained CheY-P phosphatase activity despite its conserved dimeric fold structure exhibiting divergence in active-site architecture, suggesting a preserved catalytic mechanism among distantly related homologs. Co-conservation of cheX and cheZ across disparate bacterial phyla suggests their adaptative retention confers robustness and versatility to chemotactic control. In a bacterium where they coexist, CheX tuned CheY-P–dependent motility under looming nutrient depletion, while CheZ did not, and their co-conservation likely provide robust, versatile control of stay-or-go foraging decisions across diverse bacteria.

|
Scooped by
mhryu@live.com
January 6, 4:19 PM
|
Marine diazotrophs are microscopic planktonic organisms ubiquitous in the ocean, that play a major ecological role: they supply nitrogen to the surface ocean biosphere, an essential but scarce nutrient in ~60% of the global ocean. Over the past decades, they have attracted considerable attention, with numerous studies providing key insights into their diversity, lifestyle, biogeographical distribution, and biogeochemical role in planktonic ecosystems. An increasing number of studies show that these microbes regulate marine productivity and shape the food web by alleviating nitrogen limitation, thereby contributing to carbon sequestration to the deep ocean. Yet, the diazotroph-derived organic carbon exported to the deep ocean is still poorly quantified, limiting robust estimates of the ocean’s contribution to CO₂ sequestration and climate change mitigation under present and future conditions. This knowledge gap reflects the complexity of diazotroph export pathways to the deep ocean, whose quantification and variability drivers remain difficult to resolve with current methods. This review aims to synthesize current knowledge on the role of diazotrophs in their interactions with the food web and the biological carbon pump, reanalyze existing datasets, identify key knowledge gaps, and propose future research directions.

|
Scooped by
mhryu@live.com
January 6, 4:09 PM
|
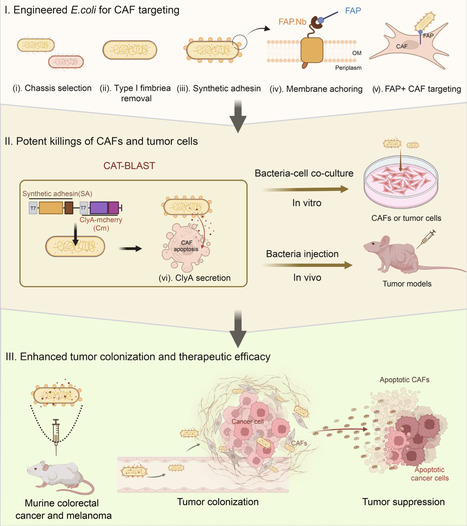
Cancer-associated fibroblasts (CAFs) construct a protective stromal barrier that promotes tumor growth and resistance to therapy. To dismantle this, we developed CAT-BLAST, an engineered bacterial platform designed to potently eliminate these cells. We engineered a safe, high-expression E. coli chassis, arming it with a FAP-specific synthetic adhesin for precise CAF targeting and the ability to secrete the ClyA cytotoxin to induce apoptosis in both CAFs and adjacent tumor cells. This platform demonstrated robust, FAP-specific targeting across diverse human tumor xenograft models and achieved significant tumor suppression in both murine colorectal cancer and melanoma.

|
Scooped by
mhryu@live.com
January 6, 4:03 PM
|
Continuous directed evolution is a powerful Synthetic Biology tool to engineer proteins with desired functions in vivo. Mimicking natural evolution, it involves repeated cycles of high-frequency mutagenesis, selection, and replication within platform cells, where the function of the target gene is tightly linked to the host cell’s fitness. However, cells might escape the selection pressure due to the inherent flexibility of their metabolism, which allows for adaptation. Whole-proteome analysis as well as targeted proteomics offer valuable insights into global and specific cellular changes. They can identify modifications in the target protein and its interactors to help understand its evolution and network integration. Using the continuous evolution of the Arabidopsis thaliana methionine synthases AtMS1 and AtMS2 as an example, we show how mass spectrometry-based proteomics was able to assess the abundance of target enzymes, identify flaws in population construction, measure methionine metabolic adaptation, and allow informed decision-making in the evolution campaign.

|
Scooped by
mhryu@live.com
January 6, 3:58 PM
|
Proteases, enzymes that play critical roles in health and disease, exert their function through the cleavage of peptide bonds. Identifying substrates that are efficiently and selectively cleaved by target proteases is essential for studying protease activity and for harnessing it in protease-activated diagnostics and therapeutics. However, the vast design space of possible substrates (c.a. 2010 amino acid combinations for a 10-mer peptide) and the limited accessibility of high-throughput activity profiling tools hinder the speed and success of substrate design. We present CleaveNet, an end-to-end AI pipeline for the design of protease substrates. Applied to matrix metalloproteinases, CleaveNet enhances the scale, tunability, and efficiency of substrate design. CleaveNet generates peptide substrates that exhibit sound biophysical properties and capture not only well-established but also previously-uncharacterized cleavage motifs. To control substrate design, CleaveNet incorporates a conditioning tag that steers peptide generation towards desired cleavage profiles, enabling targeted design of efficient and selective substrates. CleaveNet-generated substrates were validated experimentally through a large-scale in vitro screen, even in the challenging case of designing highly selective substrates for MMP13. We envision that CleaveNet will accelerate our ability to study and capitalize on protease activity, paving the way for in silico design tools across enzyme classes. Effective substrates are key to probing and harnessing protease activity. This work presents CleaveNet, an AI tool that generates efficient, selective substrates, revealing known and distinct cleavage motifs and tuning designs to target activity profiles.

|
Scooped by
mhryu@live.com
January 6, 3:40 PM
|
Enzymes are environmentally friendly biocatalysts that play a crucial role in catalyzing biochemical reactions. However, the catalytic performance of natural enzymes often cannot fully meet the demands of industrial production. Rapidly developing AI tool-assisted protein engineering modification strategies can be used to significantly enhance the catalytic properties of enzymes. The review highlights the functional features of different protein language models and their representative tools. In addition, the system reviews various software tools used for analyzing protein catalytic performance evaluation indicators and provides a detailed comparison of the success rates and application permissions of different tools. Finally, the challenges and future directions of AI techniques in protein engineering are emphasized to strengthen the role of rational computations in protein design and personalization. This review provides a comprehensive perspective for researchers to promote further development in the field of protein engineering.

|
Scooped by
mhryu@live.com
January 6, 3:33 PM
|
Sequence alignment is essential for genomic research and clinical diagnostics, yet detecting complex rearrangements such as inversions, duplications, and gene conversions remains challenging due to allele complexity and limitations of current methods. We introduce VACmap, a non-linear mapping approach to enhance the detection and representation of all genetic variations. VACmap improves duplication detection from 20% to 90% in the Challenging Medically-Relevant Genes (CMRG) benchmark and improves characterization of complex inversions in repetitive regions and gene conversion events. It improves resolving clinically significant loci, including the LPA gene (with repetitive KIV-2 units linked to coronary heart disease), GBA1 and STRC genes (risk factors for Parkinson’s disease and hearing loss, respectively, affected by pseudogene recombination with GBAP1 and STRCP1). Here, we show that VACmap delivers better alignment accuracy and SV detection, providing a robust tool for genomic analysis and clinical insights, with potential to advance understanding of genetic diversity and disease mechanisms. Here the authors introduce VACmap, a nonlinear long-read aligner that improves detection of complex structural variations like duplications, inversions, and gene conversions. It enhances SV callers’ performance on benchmarks and resolves clinically relevant loci in LPA, GBA1, and STRC genes.

|
Scooped by
mhryu@live.com
January 6, 1:00 AM
|
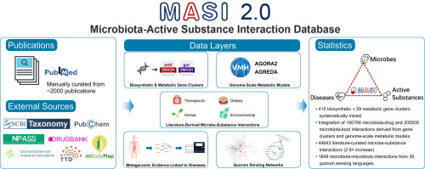
Extensive interactions between microbiota and active substances are health- and disease-relevant. Mechanistic understanding from genomic perspective of these interactions and potential impacts is important for biomedical and pharmaceutical research. However, current data repositories often lack systematic integration from a genomic perspective. Here we describe an update of the MASI microbiota-active substance interactions database. This update includes new data of (1) genomic-derived 166,766 microbiota-drug interactions and 205,505 microbiota-food interactions linked by 415 biosynthetic gene clusters (BGCs), 59 metabolic gene clusters (MGCs), and 7250 genome-scale metabolic network models (GEMs) of ∼1200 microbiota species, and (2) 1848 microbiota-microbiota interaction records mediated by 39 quorum sensing languages, and (3) 46,717 microbiota-disease associations between 640 species and 59 diseases. Overall, this update provides 44,643 interasctions derived from ∼2000 publications and 380,571 genome-derived interactions, covering 1867 microbe species, 1576 therapeutic substances, 357 dietary substances, which is freely accessible at https://www.aiddlab.com/MASI2025/index.html.
|
 Your new post is loading...
Your new post is loading...



























CRISPRi, now Csys https://www.sciencedirect.com/science/article/abs/pii/S2405471225002601