 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 11:03 PM
|
This study presents Bacillus subtilis T7 as the first known strain of B. subtilis capable of simultaneous lignin depolymerization and direct hydrogen production—a dual metabolic capability not previously reported in this species. B. subtilis T7 demonstrated 63.38% lignin degradation and 56.53% Azure B decolorization over seven days, with HPLC detection of aromatic intermediates—ferulic acid (0.85 mg/L) and vanillin (0.666 mg/L)—confirming active lignin catabolism. Agar-based assays revealed robust hydrolytic enzyme activities, including proteases (23.3 mm), cellulases (24.5 mm), amylases (14.2 mm), and xylanases (11.6 mm), surpassing those of many reported Bacillus strains. Whole-genome analysis confirmed a cascade of carbohydrate-active enzymes (CAZymes) including AA10 (lytic polysaccharide monooxygenases), AA3 (oxidoreductases), AA6 (quinone reductases), and CE1 (acetyl xylan esterases). These enzymes are associated with enhanced cellulolytic and xylanolytic activities, as well as increased lignin degradation. Batch fermentation experiments demonstrated that B. subtilis T7 produced hydrogen yields ranging from 0.53 to 1.41 mol H₂/mol substrate across various feedstocks, including xylose, glucose, carboxymethyl cellulose (CMC), starch, and untreated food waste. Xylose exhibited the highest volumetric productivity, achieving 274 mL H₂/g volatile solids, along with the most rapid production kinetics, indicating efficient metabolic utilization of this pentose sugar. In contrast, untreated food waste yielded the maximum molar hydrogen output of 1.41 mol H₂/mol substrate, attributable to its heterogeneous carbohydrate composition and lower average molecular weight, which likely enhanced enzymatic accessibility and substrate solubilization. These findings indicate that B. subtilis T7 encodes a functional [FeFe]-hydrogenase operon, along with an expanded repertoire of oxidative CAZymes, enabling it to bioprocess waste biomass into hydrogen without the need for syntrophic partners.

|
Scooped by
mhryu@live.com
Today, 10:01 PM
|
Rhizosphere microbes benefit plant growth and health. How plant-microbe interactions regulate fruit quality remains poorly understood. Here, we elucidate the multi-level modulation of vitamin accumulation in tomato by flavonoid-mediated crosstalk between host plants and rhizosphere microbes. SlMYB12-overexpressing plants with up-regulated flavonoid biosynthesis accumulate higher levels of vitamins C and B6 in fruits compared to wild-type plants grown in natural soil. Flavonoid-mediated improvement of fruit quality depends on the presence of soil microbiomes and relates to rhizosphere enrichment of key taxa (e.g. Lysobacter). Multi-omics analyses reveal that flavonoids attract Lysobacter soli by stimulating its twitching motility and spermidine biosynthesis, which in turn boosts vitamin accumulation in fruits across tomato cultivars and soil types. RpoN acts as a dual regulator in L. soli that is responsive to flavonoids, controlling bacterial motility and spermidine production. Our study provides insight into flavonoid-mediated rhizosphere signalling and underscores plant-microbiome orchestration for improved tomato fruit quality. How fruit quality is regulated by plant microbiome remains poorly understood. Here, the authors reveal that flavonoids secreted by tomato roots can recruit specific soil microbes to the rhizosphere and stimulate spermidine biosynthesis, which can induce vitamin accumulation in tomato fruits.

|
Scooped by
mhryu@live.com
Today, 9:45 PM
|
Heteroresistance is a transient resistance phenotype characterized by the presence of small subpopulations of bacterial cells with elevated antibiotic resistance within a susceptible main population. In gram-negative pathogens, heteroresistance is frequently caused by tandem amplification of genes encoding resistance proteins with low activity toward the antibiotic, a process commonly mediated by homologous recombination between flanking repeated sequences. However, the specific roles of individual recombination proteins in this mechanism remain largely undefined. In this study, we systematically evaluated the contribution of 19 recombination-associated genes to tandem amplification-driven heteroresistance in Escherichia coli. A clinical plasmid causing tobramycin heteroresistance by tandem amplification of the aac(3)-IId gene was conjugated into recombination gene-deficient mutants and the wild-type parental strain. While heteroresistance was observed with all mutants, the frequency of resistant subpopulations was decreased in recA and recB mutants, and a shift in resistance mechanism toward increased plasmid copy number and resistance mutations was observed. Partially reduced frequencies of tandem amplifications and a shift toward other heteroresistance mechanisms were also observed with recC, recJ, ruvA, and ruvC mutants, whereas other deletions of recombination genes had no or little impact on tandem amplifications. These findings identify RecABC as a key pathway in heteroresistance via tandem amplification, but even when these genes are deleted, resistant subpopulations can still be generated by other mechanisms.

|
Scooped by
mhryu@live.com
Today, 3:53 PM
|
Bacterial microcompartments (BMCs) are pseudo-organelles that sequester metabolic enzymes, intermediates, and/or gases within the bacterial cytosol. One model BMC is the carboxysome (CB). CBs facilitate rubisco-driven fixation of CO2, increasing efficiency and maximizing the phosphoglycerate output in CB-containing bacteria. The α-CBs are of particular interest due to their small size and relative simplicity, making them ideal targets for bioengineering applications. These CBs were the first BMC observed and have been a long-studied model; however, they are challenging to study in native systems and in purified samples. Recent advances in cryogenic electron microscopy and cryogenic electron tomography have resulted in many new published structures of the shell proteins, shell assemblies, and cargo organization within the CB. These new insights have advanced the field’s understanding of important structural interfaces, shed insights into once unknown domain functions, and the complex mechanisms involved in assembly and maintenance of the CB. This review highlights recently published structures of α-CB proteins and the functional and mechanistic findings of these studies.

|
Scooped by
mhryu@live.com
Today, 1:00 PM
|
Transformer models, neural networks that learn context by identifying relationships in sequential data, underpin many recent advances in artificial intelligence. Nevertheless, their inner workings are difficult to explain. Here, we find that a transformer model within the AlphaFold architecture uses simple, sparse patterns of amino acids to select protein conformations. To identify these patterns, we developed a straightforward algorithm called Conformational Attention Analysis Tool (CAAT). CAAT identifies amino acid positions that affect AlphaFold’s predictions substantially when modified. These effects are corroborated by experiments in several cases. By contrast, modifying amino acids ignored by CAAT affects AlphaFold predictions less, regardless of experimental ground truth. Our results demonstrate that CAAT successfully identifies the positions of some amino acids important for protein structure, narrowing the search space required to make effective mutations and suggesting a framework that can be applied to other transformer-based neural networks.

|
Scooped by
mhryu@live.com
Today, 10:55 AM
|
A recent breakthrough study by Zhu et al. introduced the platform GRAPE (geminivirus replicon-assisted in planta directed evolution). GRAPE remediates plant-directed evolutionary bottlenecks by linking rolling-circle replication (RCR) to protein function; selection occurs via replicon amplification, delivering microbe-like throughput in planta while preserving native plant signaling and defense.

|
Scooped by
mhryu@live.com
Today, 10:42 AM
|
Protoplast-based systems provide a powerful and versatile platform for exploring how plants sense and respond to their environment. By enabling the direct delivery of proteins, DNA, and RNA into plant cells after cell wall removal, this approach facilitates precise molecular dissection of signaling, stress adaptation, and gene regulation across both model species and economically important crops. In this review, we analyzed 1050 published articles and categorizing them by delivery methods, research focus, plant species, and tissue types. We further highlight recent advances, including the application of single-cell transcriptomics, which provides unprecedented resolution for dissecting cellular responses and offers deeper insights into the mechanisms underlying stress resilience. Importantly, protoplast regeneration is gaining renewed attention not only as a model system for studying cellular reprogramming but also as a practical platform for crop improvement. Applications of protoplast regeneration include protoplast fusion, which integrates nuclear and organellar DNA/genomes from divergent parents to accelerate breeding and enhance tolerance to both biotic and abiotic stresses. Another important application is CRISPR/Cas ribonucleoprotein (RNP)-based editing targeting stress-resilience-related genes. In asexually propagated or highly heterozygous perennial crops with limited sexual reproduction, protoplast-based RNP delivery offers a viable and regulation-compliant strategy. This approach may help address public concerns over transgenic technologies while enabling the rapid development of stress-tolerant cultivars.

|
Scooped by
mhryu@live.com
Today, 10:35 AM
|
The advancement of technology in recent decades has given us an unprecedented ability to observe the natural world. With modern sequencing and bioinformatics technologies, we can obtain more information about the microscopic world, and its interactions with the macroscopic world, than ever before. However, fungal studies that use meta'omic technologies have been sparse compared with bacterial and plant-focused studies. In this review, we highlight the ways that meta'omics can help to address pressing questions in belowground plant-fungal ecology, show consistencies that are emerging – and discrepancies that still exist – among analysis pipelines, and advocate for reporting standards that will allow meta'omic research to more fully benefit fungal ecology.

|
Scooped by
mhryu@live.com
Today, 10:25 AM
|
Genome editing has revolutionized the treatment of genetic diseases, yet the difficulty of tissue-specific delivery currently limits applications of editing technology. In this Review, we discuss preclinical and clinical advances in delivering genome editors with both established and emerging delivery mechanisms. Targeted delivery promises to considerably expand the therapeutic applicability of genome editing, moving closer to the ideal of a precise ‘magic bullet’ that safely and effectively treats diverse genetic disorders. Doudna and colleagues discuss recent advances in the targeted delivery of genome editors in vivo, offering a framework for the rational design of delivery systems.

|
Scooped by
mhryu@live.com
Today, 10:17 AM
|
The filamentous fungus Trichoderma reesei has tremendous capability to secrete proteins. Therefore, it would be an excellent host for producing high levels of therapeutic proteins at low cost. Developing a filamentous fungus to produce sensitive therapeutic proteins requires that protease secretion is drastically reduced. We have identified 13 major secreted proteases that are related to degradation of therapeutic antibodies, interferon alpha 2b, and insulin like growth factor. The major proteases observed were aspartic, glutamic, subtilisin-like, and trypsin-like proteases. The seven most problematic proteases were sequentially removed from a strain to develop it for producing therapeutic proteins. After this the protease activity in the supernatant was dramatically reduced down to 4% of the original level based upon a casein substrate. When antibody was incubated in the six protease deletion strain supernatant, the heavy chain remained fully intact and no degradation products were observed. Interferon alpha 2b and insulin like growth factor were less stable in the same supernatant, but full length proteins remained when incubated overnight, in contrast to the original strain. As additional benefits, the multiple protease deletions have led to faster strain growth and higher levels of total protein in the culture supernatant.

|
Scooped by
mhryu@live.com
Today, 1:24 AM
|
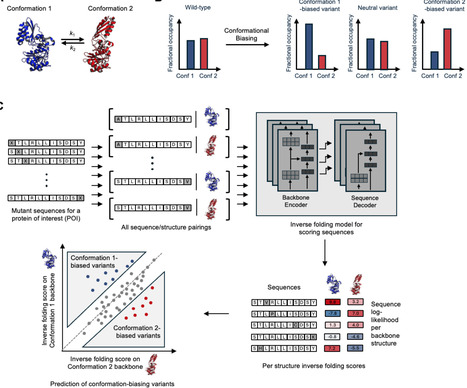
Conformational biasing (CB) is a rapid and streamlined computational method that uses contrastive scoring by inverse folding models to predict protein variants biased toward desired conformational states. We successfully validated CB across seven diverse datasets, identifying variants of K-Ras, SARS-CoV-2 spike, β2 adrenergic receptor, and Src kinase with improved conformation-specific functions, such as enhanced binding or enzymatic activity. Applying CB to the enzyme lipoic acid ligase (LplA), we uncovered a previously unknown mechanism controlling its promiscuous activity. Variants biased toward an “open” conformation state became more promiscuous, whereas “closed”-biased variants were more selective, enhancing LplA’s utility for site-specific protein labeling with fluorophores in living cells. The speed and simplicity of CB make it a versatile tool for engineering protein dynamics with broad applications in basic research, biotechnology, and medicine.

|
Scooped by
mhryu@live.com
Today, 1:08 AM
|
The dark-operative protochlorophyllide oxidoreductase (DPOR) catalyzes the light-independent reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), a key step in photosynthetic pigment biosynthesis. Structurally and mechanistically related to nitrogenase, DPOR consists of a reductase (BchL) and a catalytic component (BchNB) homologous to the reductase (NifH) and catalytic component (NifDK) of Mo-nitrogenase. Structural alignment of Rhodobacter capsulatus (Rc) BchNB with Azotobacter vinelandii (Av) NifDK and the cofactor maturase NifEN reveals a conserved α2β2 architecture and a shared cofactor-insertion path linking their respective prosthetic-like group/cofactors (Pchlide, M-cluster, L-cluster), suggesting the possibility of generating chimeric proteins with novel reactivities. Herein, Pchlide-free RcBchNB (RcBchNBapo) is reconstituted with the L-cluster extracted from AvNifEN to yield a hybrid protein (RcBchNBL) capable of reducing N2 and C1 substrates (CN−, CO) to NH3 and hydrocarbons, respectively, in the presence of a strong reductant (EuII-DTPA). In contrast, reconstituting Pchlide-bound RcBchNB with the L-cluster yields minimal activity, indicating that Pchlide and the L-cluster compete for a common binding site, as supported by Boltz-2 modeling. These findings support the hypothesis of an intertwined evolution of photosynthetic and nitrogen-fixing enzymes and outline a framework for engineering chimeric metalloenzymes that couple light capture with nitrogenase-like catalysis in the future.

|
Scooped by
mhryu@live.com
January 11, 12:18 PM
|
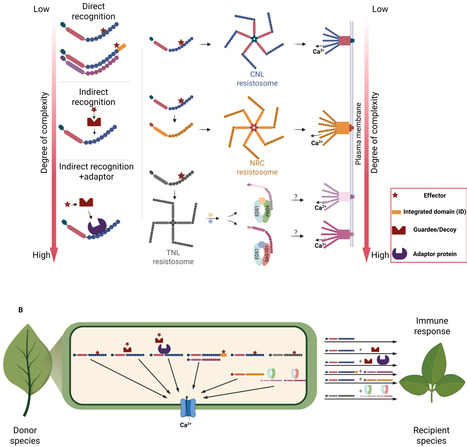
Plants employ cell-surface and intracellular immune receptors to perceive pathogens and activate defense responses. Recent advances in mechanistic understanding of how cell-surface and intracellular immune receptors convert recognition of molecular patterns or effectors into defense activation, combined with the knowledge of receptor repertoire variation both within and between species, allow transfer of immune receptors between species to increase the spectrum of recognition specificities. Here, we summarize recent progress in the functional transfer of immune receptors within and between plant families. We also discuss challenges that limit the transferability of intracellular immune receptors, including the requirement of additional host factors or downstream components and their incompatibility between donor and recipient species. Finally, we provide an overview of future perspectives for bioengineering disease-resistant crops through immune receptor transfer.
|

|
Scooped by
mhryu@live.com
Today, 10:18 PM
|
Hyperlysinemia is a life-threatening metabolic disorder that requires the continuous clearance of lysine. Engineered probiotics capable of degrading lysine in the gut represent a promising therapeutic strategy. However, the introduction of heterologous metabolic pathways can impose a substantial fitness burden on the bacterial host, potentially compromising the therapeutic efficacy. Current screening methods fail to adequately assess this pathway-induced stress. Therefore, optimizing methods to evaluate bacterial fitness after pathway modification is essential for developing effective bacterial therapies. Here, we present a label-free phenotypic screening approach using Fourier transform infrared (FTIR) spectroscopy to evaluate the physiological burden imposed by two distinct lysine catabolism pathways engineered E. coli Nissle 1917 (EcN): the plant-derived bifunctional enzyme LKR-SDR and the yeast-derived two-enzyme cascade Lys2-Lys5. Employing FTIR under lysine stress mimicking pathological concentrations, decoded pathway-specific stress signatures, and molecular resilience. Probiotics expressing LKR-SDR exhibited severe multisystem damage, including proteotoxicity, lipid peroxidation, and significant nucleic acid stress. In contrast, the Lys2-Lys5 strain demonstrated superior resilience, maintained structural integrity, and exhibited adaptive metabolic changes, primarily through lipid membrane remodeling. This study establishes FTIR spectroscopy as a rapid screening platform that identifies the Lys2-Lys5 pathway as optimal for probiotic therapies. By directly linking spectroscopic signatures to cellular fitness, FTIR spectroscopy accelerates the rational development of durable microbial therapeutics for inborn metabolic disorders.

|
Scooped by
mhryu@live.com
Today, 9:48 PM
|
Streptomyces produce a multitude of secondary metabolites, which have been exploited in drug discovery campaigns for more than three-quarters of a century. Our understanding of microbial physiology has been revolutionized by genome sequencing and large-scale functional studies. Technology for genome-wide investigations in Streptomyces species, however, has lagged behind that for other bacterial systems, hindering exploitation of unprecedented quantities of genomic data. Here, we develop a platform for en masse CRISPRi-seq for Streptomyces spp. By performing CRISPRi-seq with 2,160 unique sgRNAs targeting all operons (432 operons) encoding membrane transporters (629 genes) representing 1.1Mb of the 6.8Mb genome for S. albidoflavus, combined with hit validation, we discovered that only a small proportion (13 of 432 operons, 25 kb) contribute positively to fitness. Our work provides both a first-in-class platform for high-throughput functional genomics and a generalized blueprint for en masse screens in Streptomyces species.

|
Scooped by
mhryu@live.com
Today, 4:05 PM
|
Ammonia present in the environment is a major source of nitrogen, but it can be toxic to bacteria. While the biochemical mechanisms involved in the metabolic detoxification of cellular ammonia are well understood, little is known about how bacteria manage toxic external ammonia to survive, especially when ammonia is present as a waste product at high concentrations. Here, we demonstrate that a two-component system consisting of the sensor kinase GrtK and the response regulator GrtR is responsible for sensing and neutralizing toxic environmental ammonia produced as a waste product by the rice pathogen Burkholderia glumae. The growth of null mutants of grtK or grtR was inhibited in amino acid-rich media such as Luria-Bertani medium, but no growth inhibition was observed in amino acid-free media. The expression of obcAB, responsible for the biosynthesis of the previously known neutralizing agent oxalate, was dependent upon external ammonia concentration in a GrtR-dependent manner. Significant changes in fluorescence were observed when cells of B. glumae carrying a recombinant plasmid of the modified circular permutation GFP gene fused to grtK were incubated with compounds containing ammonium, suggesting that GrtK interacts selectively with external ammonia. Transcriptome analysis of grtK and grtR mutants also showed that GrtK and GrtR are involved in the metabolic detoxification of cellular ammonia as well. These results indicate that GrtK is an external ammonia sensor that is not a member of the ammonia transporter protein family and works together with the response regulator GrtR to counter the risk posed by its own metabolism.

|
Scooped by
mhryu@live.com
Today, 3:50 PM
|
Microbial competition serves as a fundamental driver for the evolution of offensive and defensive mechanisms among microorganisms. While it is well established that bacteria utilize specialized secretion systems to deliver both toxic and non-toxic effector proteins into competing cells, thereby directly killing them or modulating cellular events, it remains largely unclear whether bacteria can hijack effector proteins derived from competitors to resolve interspecies conflicts. Here, we demonstrate that Pseudomonas protegens employs a sophisticated defense strategy, hijacking LtaE, a non-cytotoxic effector delivered via the type IV secretion system (T4SS) of non-flagellated Lysobacter enzymogenes, to resolve interbacterial conflict through transcriptional reprogramming. Translocated LtaE neutralizes an uncharacterized antibacterial toxin in P. protegens. Surprisingly, as a countermeasure, P. protegens hijacks LtaE to rewire its host signaling hierarchy, converting it into a motility activation switch. Mechanistically, LtaE directly binds to FleQ, the σ54-dependent master regulator of flagellar biosynthesis, shielding it from inhibitory c-di-GMP binding—a universal second messenger whose elevated concentration typically inhibits bacterial motility and promotes biofilm formation. Remarkably, the LtaE-FleQ complex remains stable under high c-di-GMP conditions, overriding sessility signals to derepress flagellar gene expression and trigger escape motility. Biochemical analyses reveal that LtaE broadly targets FleQ homologs across pseudomonads through competitive inhibition of c-di-GMP binding, linking competitor detection to motility activation. Our findings establish a novel bacterial conflict-resolution paradigm, demonstrating how non-cytotoxic effectors act as molecular switches to dynamically reprogram transcriptional networks and enhance phenotypic plasticity.

|
Scooped by
mhryu@live.com
Today, 11:55 AM
|
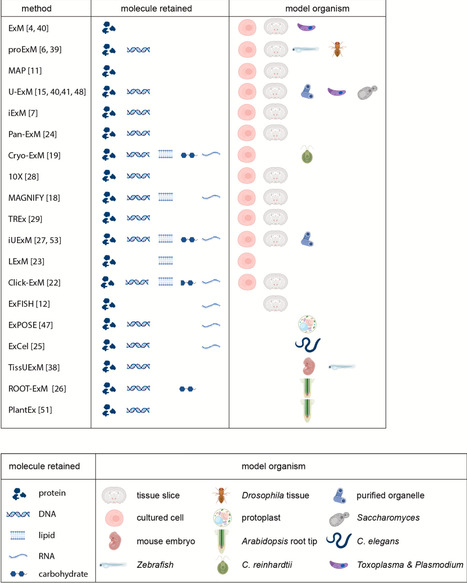
Super-resolution microscopy (SRM) has transformed the study of cellular structures, enabling imaging beyond the diffraction limit. Yet, the need for costly instrumentation has limited its accessibility. Expansion Microscopy microscopy (ExM), pioneered by Ed Boyden’s laboratory, offers an alternative by physically enlarging samples embedded within a swellable hydrogel. This simple principle makes nanoscale resolution achievable with conventional fluorescence microscopy. Since its introduction in 2015, ExM has rapidly diversified. Iterative ExM (iExM) increases resolution through repetitive expansion, chemical gel innovation enables single-step 10- to 20-fold expansion, and hybrid strategies combining ExM and SRM techniques have pushed resolution below 15 nm. ExM has now been applied to diverse biological models but its adaptation to complex plant tissues poses unique challenges due to their rigid cell walls. Recent advances in the field of plant science have started to address these obstacles, opening access to nanoscale imaging of plant cellular structures such as plasmodesmata and the mitotic spindle. In this review, we trace the development of ExM from its pioneering stages to current refinements, discuss methodological advances and hybrid approaches, examine technical limitations, and highlight emerging applications across biological models, with a particular focus on recent progress and future perspectives in plant biology.

|
Scooped by
mhryu@live.com
Today, 10:46 AM
|
Microorganisms inhabit diverse environments, including nearly every organ in the human body. The human microbiome—a complex community of microorganisms residing in the human body—has gained increasing attention as a key contributor to human health and disease, making it an important target for the development of diagnostic and therapeutic strategies. However, the inherent complexity of microbial communities and the challenges of engineering diverse non-model microorganisms present significant barriers. To address these challenges, synthetic biology has provided powerful tools and strategies to engineer microorganisms capable of sensing disease-specific environments and performing targeted therapeutic functions. In particular, the development of synthetic genetic circuits has significantly improved the precision and reliability of disease diagnosis and treatment, enabling real-time disease monitoring, therapeutic, and even preventive interventions. This review highlights state-of-the-art synthetic biology tools and strategies for engineering the probiotics and commensal bacteria aimed at the diagnosis and treatment of human diseases, with accompanying examples. Future challenges and prospects are also discussed.

|
Scooped by
mhryu@live.com
Today, 10:40 AM
|
With more than 833 million hectares of saline soils worldwide, which are found primarily in arid and semiarid regions, soil salinization poses a serious threat to global food security and agricultural development. Therefore, a variety of saline soil improvement strategies, including physical, chemical, hydrological and biological methods, have been developed. These methods offer the possibility of rehabilitating saline-alkaline land by improving soil properties, reducing salt concentrations, and promoting plant growth. Among them, the use of microorganisms is considered to have great potential. The microbial communities in saline–alkaline soils are complex and diverse and include saline-tolerant and salinophilic microorganisms that adapt to high-salt and high-alkaline environments by regulating gene expression and altering metabolism and osmotic balance; these microorganisms play important roles in balancing soil ecosystems and maintaining soil fertility. In addition, plant growth-promoting rhizobacteria (PGPR) can increase plant tolerance to saline and alkaline stress and promote plant growth through various mechanisms, such as phytohormone modulation, osmoregulation, antioxidant defence enhancement, and improved nutrient availability. This review systematically summarizes the current understanding of these microbial adaptation mechanisms and their probiotic effects, highlighting the potential for developing efficient microbial fertilizers for the sustainable use of saline soils and the enhancement of agricultural production.

|
Scooped by
mhryu@live.com
Today, 10:33 AM
|
The presence of intracellular microbiota in epithelial cells of gastrointestinal tracts (GITs) of dairy cows, as well as their associations with rumen development, remains unclear. Using a single-cell analysis of host-microbiome interactions (SAHMI) within a single-cell atlas derived from ten GITs tissue types collected from new-born (NB) and adult (AD) cows, we found that 20.5% of the single-cell RNA sequencing reads aligned to reference microbial genomes after filtering low-quality single cells and doublets. Comparative analysis revealed that abomasum tissue exhibited the highest proportion of cells detected microbial signals, with Paneth cells possessing the most genes classified as both marker genes and those related to microbial signals. In the NB rumen, Basal cells demonstrated the greatest overlap between differentially expressed genes in AD vs. NB comparison and the microbial signal-related genes. Notably, these microbiota-associated genes, which are mainly linked to Aliiroseovarius crassostreae, Enterobacter sp. T2, and Enzebya pacifica, are implicated in nucleotide excision repair mechanisms, including DNA replication and the cell cycle. Furthermore, bacterial fluorescence in situ hybridization (FISH) analysis indicated that these three microbial species were partially localized within the cytoplasm and nucleus of rumen epithelial cells in NB cattle. These findings provide substantial evidence supporting the existence of an intracellular microbiome within the GITs of dairy cattle and highlight their potential relationships with rumen development. This research enhances our understanding of the crosstalk between hosts and microbiome during the maturation of ruminants.

|
Scooped by
mhryu@live.com
Today, 10:21 AM
|
The intestinal epithelium relies on continuous stem cell-driven renewal to maintain barrier function and recover from injury. While bacterial signals are known to influence intestinal stem cell behavior, the regenerative capacity of the gut mycobiome has remained largely unexplored. Here we identify the commensal fungus Kazachstania pintolopesii (Kp) as a critical mediator of intestinal regeneration through its secreted protein Ygp1. We found that a 12-amino acid peptide fragment of Ygp1, CD12, was sufficient to promote intestinal organoid differentiation and accelerate intestinal healing in murine models of colitis and chemotherapy-induced injury. Transcriptomics, simulations and molecular interaction experiments revealed that CD12 binds mammalian α-enolase (ENO1), enhancing YAP1 (Yes-associated protein 1) protein levels and activating regenerative transcriptional programmes through the Hippo signalling pathway. Engineered probiotics expressing CD12 replicated its therapeutic benefits, offering a translatable delivery strategy. Our work expands the therapeutic potential of the mycobiome, positioning it as a source of biologics for inflammatory and iatrogenic gut disorders. A short peptide derived from a commensal-fungal-secreted protein promotes repair in models of colon epithelial damage when delivered directly or via probiotic microbes.

|
Scooped by
mhryu@live.com
Today, 10:01 AM
|
Prime editing (PE) enables precise genome modifications without donor DNA or double-strand breaks, but its application in dicot plants has faced challenges due to low efficiency, locus dependence, and poor heritability. Here, we develop an ultra-efficient prime editing (UtPE) system for dicots by integrating evolved PE6 variants (PE6c and PE6ec), an altered pegRNA (aepegRNA), an RNA chaperone, and a geminiviral replicon. UtPE significantly improves editing performance in tomatoes, with UtPEv1 excelling in simple edits (unstructured RTTs) and UtPEv3 effective for complex targets (structured RTTs or multiple nucleotide changes). Compared to a PE2max-based tool, UtPE increases desired editing efficiency by 3.39 to 8.89-fold, enables editing at previous inaccessible sites, achieves an average of 16.0% desired editing efficiency in calli, and produces high-frequency desired edits in up to 87.5% of T0 plants. Multiplexed editing at up to three loci and stable T1 inheritance are also achieved, resulting in traits such as jointless pedicels and glyphosate resistance, while minimizing off-target effects. Low efficiency, locus-dependence, and poor heritability affect the application of prime editing to dicot plants. Here, the authors solve these problems by developing an ultra-efficient prime editing (UtPE) system for dicots by integrating evolved PE6 variants, an altered pegRNA, an RNA chaperone, and a geminiviral replicon.

|
Scooped by
mhryu@live.com
Today, 1:12 AM
|
Improving the photosynthetic enzyme Rubisco is a key target for enhancing C3 crop productivity, but progress has been hampered by the difficulty of evaluating engineered variants in planta without interference from the native enzyme. Here, we report the creation of a Rubisco-null plant Nicotiana tabacum platform by using CRISPR-Cas9 to knock out all 11 nuclear-encoded small subunit (rbcS) genes. Knockout was achieved in a line expressing cyanobacterial Rubisco from the plastid genome, allowing the recovery of viable plants. We then developed a chloroplast expression system for coexpressing both large and small subunits from the plastid genome. We expressed two resurrected ancestral Rubiscos from the Solanaceae family. The resulting transgenic plants were phenotypically normal and accumulated Rubisco to wild-type levels. Importantly, kinetic analyses of the purified ancestral enzymes revealed they possessed a 16 to 20% higher catalytic efficiency (kcat,air/Kc,air) under ambient conditions, driven by a significantly faster turnover rate (kcat,air). We have demonstrated that our system allows robust in vivo assessment of novel Rubiscos and that ancestral reconstruction is a powerful strategy for identifying superior enzymes to improve photosynthesis in C3 crops.

|
Scooped by
mhryu@live.com
January 11, 11:19 PM
|
Soil biodiversity is a key driver of ecosystem function, including nutrient cycling, organic-matter decomposition, plant productivity, climate regulation and pathogen control (with subsequent effects on animal, human and plant health). A foundational review in 2014 described the functional role of soil biodiversity in ecosystems, but our understanding of the relationship between soil biodiversity and ecosystem functioning has deepened over the past decade. In this Review, we highlight progress in the field, discuss the approaches and methodological advances that have enabled this progress, and identify emerging research questions. Although the spatiotemporal patterns and community dynamics of soil communities are becoming well understood, topics with important knowledge gaps include the climate feedback effects of soils, the ecology of urban soils and the development of soil health indicators. Global collaborative networks, linking existing databases, and monitoring soil biodiversity and ecosystem functioning are important ways to address these knowledge gaps. By considering the relationships between soil biodiversity and ecosystem functioning we can connect small-scale interactions among plants, microorganisms and animals to ecosystem services and planetary sustainability. Soil biodiversity is poorly studied compared to aboveground biodiversity, but is an important driver of ecosystem functioning. This Review discusses advances in research into the relationships between soil biodiversity and ecosystem functioning, highlighting that integrative and causal study approaches will be needed to fill the gaps in our understanding.
|
 Your new post is loading...
Your new post is loading...































DogTag? In the presence of a multivalent target, the binders bind to the target, resulting in a spatial separation of DarkBiT and LgBiT, provided the intramolecular affinities and linkers allow the conformational change. This allows SmBiT to complement LgBiT and form an active luciferase complex that generates light by conversion of furimazine. hibit