Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 11:55 AM
|
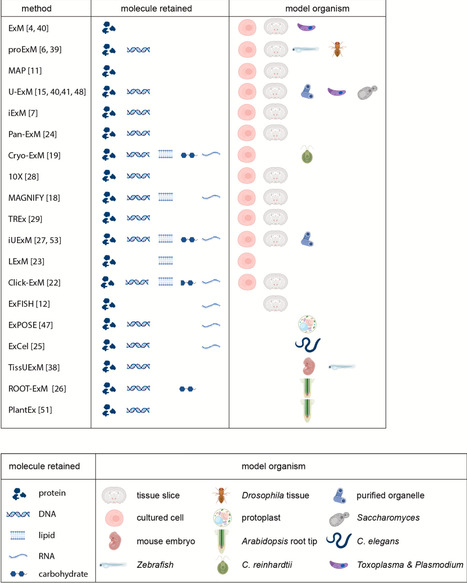
Super-resolution microscopy (SRM) has transformed the study of cellular structures, enabling imaging beyond the diffraction limit. Yet, the need for costly instrumentation has limited its accessibility. Expansion Microscopy microscopy (ExM), pioneered by Ed Boyden’s laboratory, offers an alternative by physically enlarging samples embedded within a swellable hydrogel. This simple principle makes nanoscale resolution achievable with conventional fluorescence microscopy. Since its introduction in 2015, ExM has rapidly diversified. Iterative ExM (iExM) increases resolution through repetitive expansion, chemical gel innovation enables single-step 10- to 20-fold expansion, and hybrid strategies combining ExM and SRM techniques have pushed resolution below 15 nm. ExM has now been applied to diverse biological models but its adaptation to complex plant tissues poses unique challenges due to their rigid cell walls. Recent advances in the field of plant science have started to address these obstacles, opening access to nanoscale imaging of plant cellular structures such as plasmodesmata and the mitotic spindle. In this review, we trace the development of ExM from its pioneering stages to current refinements, discuss methodological advances and hybrid approaches, examine technical limitations, and highlight emerging applications across biological models, with a particular focus on recent progress and future perspectives in plant biology.

|
Scooped by
mhryu@live.com
Today, 10:46 AM
|
Microorganisms inhabit diverse environments, including nearly every organ in the human body. The human microbiome—a complex community of microorganisms residing in the human body—has gained increasing attention as a key contributor to human health and disease, making it an important target for the development of diagnostic and therapeutic strategies. However, the inherent complexity of microbial communities and the challenges of engineering diverse non-model microorganisms present significant barriers. To address these challenges, synthetic biology has provided powerful tools and strategies to engineer microorganisms capable of sensing disease-specific environments and performing targeted therapeutic functions. In particular, the development of synthetic genetic circuits has significantly improved the precision and reliability of disease diagnosis and treatment, enabling real-time disease monitoring, therapeutic, and even preventive interventions. This review highlights state-of-the-art synthetic biology tools and strategies for engineering the probiotics and commensal bacteria aimed at the diagnosis and treatment of human diseases, with accompanying examples. Future challenges and prospects are also discussed.

|
Scooped by
mhryu@live.com
Today, 10:40 AM
|
With more than 833 million hectares of saline soils worldwide, which are found primarily in arid and semiarid regions, soil salinization poses a serious threat to global food security and agricultural development. Therefore, a variety of saline soil improvement strategies, including physical, chemical, hydrological and biological methods, have been developed. These methods offer the possibility of rehabilitating saline-alkaline land by improving soil properties, reducing salt concentrations, and promoting plant growth. Among them, the use of microorganisms is considered to have great potential. The microbial communities in saline–alkaline soils are complex and diverse and include saline-tolerant and salinophilic microorganisms that adapt to high-salt and high-alkaline environments by regulating gene expression and altering metabolism and osmotic balance; these microorganisms play important roles in balancing soil ecosystems and maintaining soil fertility. In addition, plant growth-promoting rhizobacteria (PGPR) can increase plant tolerance to saline and alkaline stress and promote plant growth through various mechanisms, such as phytohormone modulation, osmoregulation, antioxidant defence enhancement, and improved nutrient availability. This review systematically summarizes the current understanding of these microbial adaptation mechanisms and their probiotic effects, highlighting the potential for developing efficient microbial fertilizers for the sustainable use of saline soils and the enhancement of agricultural production.

|
Scooped by
mhryu@live.com
Today, 10:33 AM
|
The presence of intracellular microbiota in epithelial cells of gastrointestinal tracts (GITs) of dairy cows, as well as their associations with rumen development, remains unclear. Using a single-cell analysis of host-microbiome interactions (SAHMI) within a single-cell atlas derived from ten GITs tissue types collected from new-born (NB) and adult (AD) cows, we found that 20.5% of the single-cell RNA sequencing reads aligned to reference microbial genomes after filtering low-quality single cells and doublets. Comparative analysis revealed that abomasum tissue exhibited the highest proportion of cells detected microbial signals, with Paneth cells possessing the most genes classified as both marker genes and those related to microbial signals. In the NB rumen, Basal cells demonstrated the greatest overlap between differentially expressed genes in AD vs. NB comparison and the microbial signal-related genes. Notably, these microbiota-associated genes, which are mainly linked to Aliiroseovarius crassostreae, Enterobacter sp. T2, and Enzebya pacifica, are implicated in nucleotide excision repair mechanisms, including DNA replication and the cell cycle. Furthermore, bacterial fluorescence in situ hybridization (FISH) analysis indicated that these three microbial species were partially localized within the cytoplasm and nucleus of rumen epithelial cells in NB cattle. These findings provide substantial evidence supporting the existence of an intracellular microbiome within the GITs of dairy cattle and highlight their potential relationships with rumen development. This research enhances our understanding of the crosstalk between hosts and microbiome during the maturation of ruminants.

|
Scooped by
mhryu@live.com
Today, 10:21 AM
|
The intestinal epithelium relies on continuous stem cell-driven renewal to maintain barrier function and recover from injury. While bacterial signals are known to influence intestinal stem cell behavior, the regenerative capacity of the gut mycobiome has remained largely unexplored. Here we identify the commensal fungus Kazachstania pintolopesii (Kp) as a critical mediator of intestinal regeneration through its secreted protein Ygp1. We found that a 12-amino acid peptide fragment of Ygp1, CD12, was sufficient to promote intestinal organoid differentiation and accelerate intestinal healing in murine models of colitis and chemotherapy-induced injury. Transcriptomics, simulations and molecular interaction experiments revealed that CD12 binds mammalian α-enolase (ENO1), enhancing YAP1 (Yes-associated protein 1) protein levels and activating regenerative transcriptional programmes through the Hippo signalling pathway. Engineered probiotics expressing CD12 replicated its therapeutic benefits, offering a translatable delivery strategy. Our work expands the therapeutic potential of the mycobiome, positioning it as a source of biologics for inflammatory and iatrogenic gut disorders. A short peptide derived from a commensal-fungal-secreted protein promotes repair in models of colon epithelial damage when delivered directly or via probiotic microbes.

|
Scooped by
mhryu@live.com
Today, 10:01 AM
|
Prime editing (PE) enables precise genome modifications without donor DNA or double-strand breaks, but its application in dicot plants has faced challenges due to low efficiency, locus dependence, and poor heritability. Here, we develop an ultra-efficient prime editing (UtPE) system for dicots by integrating evolved PE6 variants (PE6c and PE6ec), an altered pegRNA (aepegRNA), an RNA chaperone, and a geminiviral replicon. UtPE significantly improves editing performance in tomatoes, with UtPEv1 excelling in simple edits (unstructured RTTs) and UtPEv3 effective for complex targets (structured RTTs or multiple nucleotide changes). Compared to a PE2max-based tool, UtPE increases desired editing efficiency by 3.39 to 8.89-fold, enables editing at previous inaccessible sites, achieves an average of 16.0% desired editing efficiency in calli, and produces high-frequency desired edits in up to 87.5% of T0 plants. Multiplexed editing at up to three loci and stable T1 inheritance are also achieved, resulting in traits such as jointless pedicels and glyphosate resistance, while minimizing off-target effects. Low efficiency, locus-dependence, and poor heritability affect the application of prime editing to dicot plants. Here, the authors solve these problems by developing an ultra-efficient prime editing (UtPE) system for dicots by integrating evolved PE6 variants, an altered pegRNA, an RNA chaperone, and a geminiviral replicon.

|
Scooped by
mhryu@live.com
Today, 1:12 AM
|
Improving the photosynthetic enzyme Rubisco is a key target for enhancing C3 crop productivity, but progress has been hampered by the difficulty of evaluating engineered variants in planta without interference from the native enzyme. Here, we report the creation of a Rubisco-null plant Nicotiana tabacum platform by using CRISPR-Cas9 to knock out all 11 nuclear-encoded small subunit (rbcS) genes. Knockout was achieved in a line expressing cyanobacterial Rubisco from the plastid genome, allowing the recovery of viable plants. We then developed a chloroplast expression system for coexpressing both large and small subunits from the plastid genome. We expressed two resurrected ancestral Rubiscos from the Solanaceae family. The resulting transgenic plants were phenotypically normal and accumulated Rubisco to wild-type levels. Importantly, kinetic analyses of the purified ancestral enzymes revealed they possessed a 16 to 20% higher catalytic efficiency (kcat,air/Kc,air) under ambient conditions, driven by a significantly faster turnover rate (kcat,air). We have demonstrated that our system allows robust in vivo assessment of novel Rubiscos and that ancestral reconstruction is a powerful strategy for identifying superior enzymes to improve photosynthesis in C3 crops.

|
Scooped by
mhryu@live.com
January 11, 11:19 PM
|
Soil biodiversity is a key driver of ecosystem function, including nutrient cycling, organic-matter decomposition, plant productivity, climate regulation and pathogen control (with subsequent effects on animal, human and plant health). A foundational review in 2014 described the functional role of soil biodiversity in ecosystems, but our understanding of the relationship between soil biodiversity and ecosystem functioning has deepened over the past decade. In this Review, we highlight progress in the field, discuss the approaches and methodological advances that have enabled this progress, and identify emerging research questions. Although the spatiotemporal patterns and community dynamics of soil communities are becoming well understood, topics with important knowledge gaps include the climate feedback effects of soils, the ecology of urban soils and the development of soil health indicators. Global collaborative networks, linking existing databases, and monitoring soil biodiversity and ecosystem functioning are important ways to address these knowledge gaps. By considering the relationships between soil biodiversity and ecosystem functioning we can connect small-scale interactions among plants, microorganisms and animals to ecosystem services and planetary sustainability. Soil biodiversity is poorly studied compared to aboveground biodiversity, but is an important driver of ecosystem functioning. This Review discusses advances in research into the relationships between soil biodiversity and ecosystem functioning, highlighting that integrative and causal study approaches will be needed to fill the gaps in our understanding.

|
Scooped by
mhryu@live.com
January 11, 12:15 PM
|
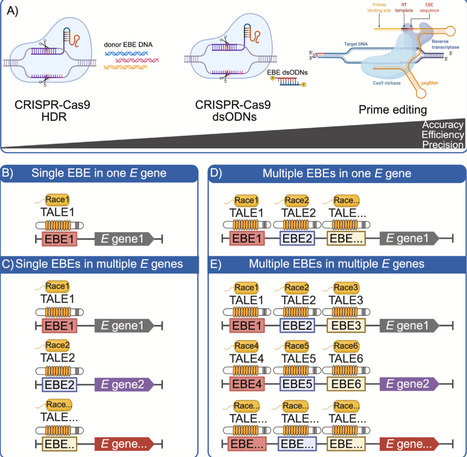
Bacterial type III effector proteins, particularly transcription activator-like effectors (TALEs) secreted by Xanthomonas spp., play critical roles in pathogen–host dynamics. While TALEs facilitate bacterial infections, they also possess vulnerabilities that plants and scientists can exploit to develop mechanisms of resistance. This review encompasses the characteristics and functions of TALEs, examining both their virulence and avirulence roles, and the host plants’ counter-strategies. We highlight advancements in genome editing technologies aimed at combating TALE-dependent plant diseases, with a focus on bacterial blight and leaf streak of rice, but also including bacterial blights of cotton and cassava, and citrus canker. Additionally, we share perspectives on various strategies and approaches for applying genome editing tools to improve disease resistance traits in crop breeding.

|
Scooped by
mhryu@live.com
January 11, 12:02 PM
|
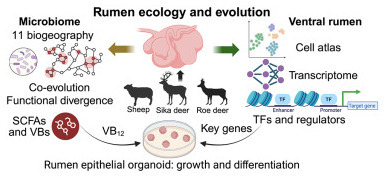
Ruminants thrive in diverse ecosystems by leveraging their rumen microbiome to ferment fibrous plants. However, the spatial biogeography of rumen microbiome and the genetic diversity of the ventral rumen epithelium remain unknown. Here, we present a multi-omics study in roe deer, sika deer, and sheep, integrating region-resolved microbiome and metabolome across 11 ruminal sacs, as well as single-cell RNA sequencing (scRNA-seq), assay for transposase-accessible chromatin using sequencing (ATAC-seq), and bulk RNA sequencing (RNA-seq) of ventral epithelium. We reveal species-specific microbial compositions and metabolic capacities contributing to differences in short-chain fatty acid and vitamin B production. We uncover functional divergence, genomic specialization, and metabolic changes across the microbiome of distinct ruminal sacs. Single-cell profiling reveals changes of immune responses and structural remodeling of the ruminal ventral epithelium. We demonstrate that vitamin B12 promotes epithelial growth and we identify genes enhancing stem cell differentiation. Our results highlight variation in microbial ecology and epithelial architecture among three ruminant species, offering insights to improve livestock productivity.

|
Scooped by
mhryu@live.com
January 11, 11:49 AM
|
Dairy industry generates significant effluent streams, comprising wastewaters and nutrient-rich by-products such as cheese whey, which pose environmental challenges for disposal, but also offer valorisation opportunities. Microalgae cultivation on whey represents a sustainable biotechnological approach that couples biomass production with effluent treatment. This study aimed at optimizing microalgal cultivation using ricotta cheese whey as substrate. Eighteen microalgal strains, mainly belonging to Scenedesmus and Chlorella, were initially screened for their ability to grow on whey. Selected strains were further tested in 300-mL photobioreactors under different management options: Low Whey Load (LWL), High Whey Load (HWL), Salinity and Micronutrient (S&M), and Moderate Whey Load (MWL) trials. Results showed that most of the tested strains grew better when cultivated on diluted whey than on standard synthetic media, with increases from 56 to 590% during the first days, and with high removal efficiencies. Several issues were identified during the trials, including nutrient shortage, salinity increase, micronutrients deficiency and potential inhibitory effects of whey compounds, which have hindered long term cultivation of microalgae on whey. In MWL trial, optimization of whey concentration (12% v/v), replenishment strategy (30% of the culture volume every 3-days), and micronutrient supplementation overcame these issues extending the cultivation period and improving biomass yield (203.6 billion cells L-1 whey-1 with the best strain). Overall, this work underscores the importance of carefully optimize microalgal cultivation management to maximise exploitation of dairy by-products and minimize growth-limiting effects of whey accumulation, thus offering a promising starting point for large-scale applications of whey in microalgae cultivation.

|
Scooped by
mhryu@live.com
January 11, 11:27 AM
|
Recent advances in high-throughput sequencing, imaging, and phenotyping have carried plant science into the era of ‘big data.’ Complex, multi-scale datasets provide new opportunities to uncover plant molecular mechanisms with a level of detail previously unachievable. Fully exploiting this complexity requires integrating advanced statistics, computational modeling, and artificial intelligence (AI). This mini-review offers guidance on how the combination of AI and mechanistic models is transforming temporal, image-based, and spatial omics data into detailed predictions of robust plant traits. In addition, embedding physical principles into AI models can enhance interpretability and strengthen their biological grounding, leading to more realistic representations of plant inner workings. Together, these advances are reshaping plant science by turning ‘big data’ into deep insights, thus greatly enriching our understanding of plant growth, adaptation, and environmental responses.

|
Scooped by
mhryu@live.com
January 10, 4:43 PM
|
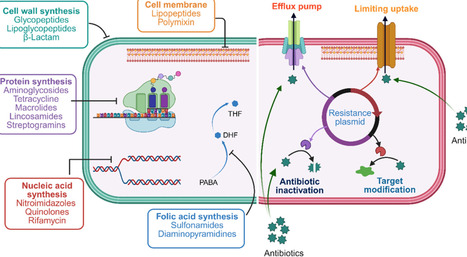
Antibiotics have revolutionized human health by significantly reducing morbidity and mortality associated with bacterial infections. Antibiotics exert bactericidal or bacteriostatic effects through inhibition of cell wall synthesis and disruption of cell membrane integrity, inhibition of protein, nucleic acid synthesis, and other metabolic pathways. Despite their remarkable success since the mid-20th century, antimicrobial resistance (AMR) has emerged as a major global health concern, undermining current treatments and complicating infection management. Key drivers of AMR include the overuse and misuse of antibiotics in clinical settings as well as bacterial adaptations such as genetic mutations and horizontal gene transfer. Mechanistically, these changes can lead to enzymatic inactivation of antibiotics, modification of drug targets, changes in permeability, and active efflux of antimicrobial agents. As resistance rises, antibiotic discovery and development have lagged, creating an urgent need for novel therapeutic strategies and chemical scaffolds. This review examines the antibiotic mechanisms and antibiotic evasion strategies, highlighting genetic and omics approaches used to identify high-priority targets for future drug discovery.
|

|
Scooped by
mhryu@live.com
Today, 10:55 AM
|
A recent breakthrough study by Zhu et al. introduced the platform GRAPE (geminivirus replicon-assisted in planta directed evolution). GRAPE remediates plant-directed evolutionary bottlenecks by linking rolling-circle replication (RCR) to protein function; selection occurs via replicon amplification, delivering microbe-like throughput in planta while preserving native plant signaling and defense.

|
Scooped by
mhryu@live.com
Today, 10:42 AM
|
Protoplast-based systems provide a powerful and versatile platform for exploring how plants sense and respond to their environment. By enabling the direct delivery of proteins, DNA, and RNA into plant cells after cell wall removal, this approach facilitates precise molecular dissection of signaling, stress adaptation, and gene regulation across both model species and economically important crops. In this review, we analyzed 1050 published articles and categorizing them by delivery methods, research focus, plant species, and tissue types. We further highlight recent advances, including the application of single-cell transcriptomics, which provides unprecedented resolution for dissecting cellular responses and offers deeper insights into the mechanisms underlying stress resilience. Importantly, protoplast regeneration is gaining renewed attention not only as a model system for studying cellular reprogramming but also as a practical platform for crop improvement. Applications of protoplast regeneration include protoplast fusion, which integrates nuclear and organellar DNA/genomes from divergent parents to accelerate breeding and enhance tolerance to both biotic and abiotic stresses. Another important application is CRISPR/Cas ribonucleoprotein (RNP)-based editing targeting stress-resilience-related genes. In asexually propagated or highly heterozygous perennial crops with limited sexual reproduction, protoplast-based RNP delivery offers a viable and regulation-compliant strategy. This approach may help address public concerns over transgenic technologies while enabling the rapid development of stress-tolerant cultivars.

|
Scooped by
mhryu@live.com
Today, 10:35 AM
|
The advancement of technology in recent decades has given us an unprecedented ability to observe the natural world. With modern sequencing and bioinformatics technologies, we can obtain more information about the microscopic world, and its interactions with the macroscopic world, than ever before. However, fungal studies that use meta'omic technologies have been sparse compared with bacterial and plant-focused studies. In this review, we highlight the ways that meta'omics can help to address pressing questions in belowground plant-fungal ecology, show consistencies that are emerging – and discrepancies that still exist – among analysis pipelines, and advocate for reporting standards that will allow meta'omic research to more fully benefit fungal ecology.

|
Scooped by
mhryu@live.com
Today, 10:25 AM
|
Genome editing has revolutionized the treatment of genetic diseases, yet the difficulty of tissue-specific delivery currently limits applications of editing technology. In this Review, we discuss preclinical and clinical advances in delivering genome editors with both established and emerging delivery mechanisms. Targeted delivery promises to considerably expand the therapeutic applicability of genome editing, moving closer to the ideal of a precise ‘magic bullet’ that safely and effectively treats diverse genetic disorders. Doudna and colleagues discuss recent advances in the targeted delivery of genome editors in vivo, offering a framework for the rational design of delivery systems.

|
Scooped by
mhryu@live.com
Today, 10:17 AM
|
The filamentous fungus Trichoderma reesei has tremendous capability to secrete proteins. Therefore, it would be an excellent host for producing high levels of therapeutic proteins at low cost. Developing a filamentous fungus to produce sensitive therapeutic proteins requires that protease secretion is drastically reduced. We have identified 13 major secreted proteases that are related to degradation of therapeutic antibodies, interferon alpha 2b, and insulin like growth factor. The major proteases observed were aspartic, glutamic, subtilisin-like, and trypsin-like proteases. The seven most problematic proteases were sequentially removed from a strain to develop it for producing therapeutic proteins. After this the protease activity in the supernatant was dramatically reduced down to 4% of the original level based upon a casein substrate. When antibody was incubated in the six protease deletion strain supernatant, the heavy chain remained fully intact and no degradation products were observed. Interferon alpha 2b and insulin like growth factor were less stable in the same supernatant, but full length proteins remained when incubated overnight, in contrast to the original strain. As additional benefits, the multiple protease deletions have led to faster strain growth and higher levels of total protein in the culture supernatant.

|
Scooped by
mhryu@live.com
Today, 1:24 AM
|
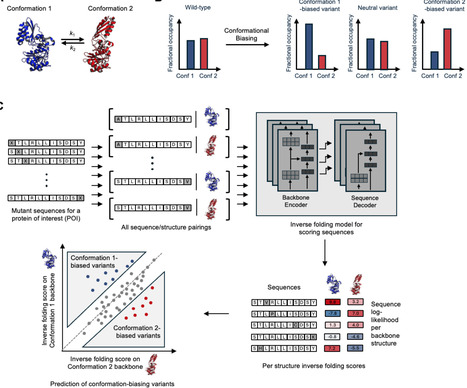
Conformational biasing (CB) is a rapid and streamlined computational method that uses contrastive scoring by inverse folding models to predict protein variants biased toward desired conformational states. We successfully validated CB across seven diverse datasets, identifying variants of K-Ras, SARS-CoV-2 spike, β2 adrenergic receptor, and Src kinase with improved conformation-specific functions, such as enhanced binding or enzymatic activity. Applying CB to the enzyme lipoic acid ligase (LplA), we uncovered a previously unknown mechanism controlling its promiscuous activity. Variants biased toward an “open” conformation state became more promiscuous, whereas “closed”-biased variants were more selective, enhancing LplA’s utility for site-specific protein labeling with fluorophores in living cells. The speed and simplicity of CB make it a versatile tool for engineering protein dynamics with broad applications in basic research, biotechnology, and medicine.

|
Scooped by
mhryu@live.com
Today, 1:08 AM
|
The dark-operative protochlorophyllide oxidoreductase (DPOR) catalyzes the light-independent reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), a key step in photosynthetic pigment biosynthesis. Structurally and mechanistically related to nitrogenase, DPOR consists of a reductase (BchL) and a catalytic component (BchNB) homologous to the reductase (NifH) and catalytic component (NifDK) of Mo-nitrogenase. Structural alignment of Rhodobacter capsulatus (Rc) BchNB with Azotobacter vinelandii (Av) NifDK and the cofactor maturase NifEN reveals a conserved α2β2 architecture and a shared cofactor-insertion path linking their respective prosthetic-like group/cofactors (Pchlide, M-cluster, L-cluster), suggesting the possibility of generating chimeric proteins with novel reactivities. Herein, Pchlide-free RcBchNB (RcBchNBapo) is reconstituted with the L-cluster extracted from AvNifEN to yield a hybrid protein (RcBchNBL) capable of reducing N2 and C1 substrates (CN−, CO) to NH3 and hydrocarbons, respectively, in the presence of a strong reductant (EuII-DTPA). In contrast, reconstituting Pchlide-bound RcBchNB with the L-cluster yields minimal activity, indicating that Pchlide and the L-cluster compete for a common binding site, as supported by Boltz-2 modeling. These findings support the hypothesis of an intertwined evolution of photosynthetic and nitrogen-fixing enzymes and outline a framework for engineering chimeric metalloenzymes that couple light capture with nitrogenase-like catalysis in the future.

|
Scooped by
mhryu@live.com
January 11, 12:18 PM
|
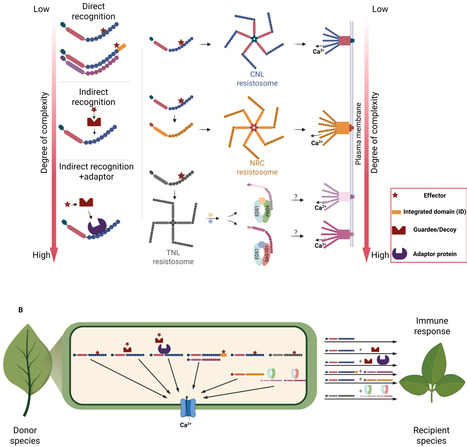
Plants employ cell-surface and intracellular immune receptors to perceive pathogens and activate defense responses. Recent advances in mechanistic understanding of how cell-surface and intracellular immune receptors convert recognition of molecular patterns or effectors into defense activation, combined with the knowledge of receptor repertoire variation both within and between species, allow transfer of immune receptors between species to increase the spectrum of recognition specificities. Here, we summarize recent progress in the functional transfer of immune receptors within and between plant families. We also discuss challenges that limit the transferability of intracellular immune receptors, including the requirement of additional host factors or downstream components and their incompatibility between donor and recipient species. Finally, we provide an overview of future perspectives for bioengineering disease-resistant crops through immune receptor transfer.

|
Scooped by
mhryu@live.com
January 11, 12:08 PM
|
The human gut microbiota, particularly the intestinal microbiota, shapes host physiology, disease risk, and therapeutic outcomes through complex metabolic and enzymatic activities. Recent advances in molecular omics, metabolomics, enzyme bioinformatics, and artificial intelligence (AI) have created unprecedented opportunities to elucidate its therapeutic roles to further enable precision microbiome medicine for personalized prevention, diagnosis, and treatment. In this review, we highlight emerging applications that leverage molecular omics and metabolomics technologies to dissect gut microbial functions, along with developments in enzyme bioinformatics and AI tools that reveal gut microbial species, enzymes, and metabolic pathways impacting human health. Finally, we discuss perspectives on data standardization, functional annotation, and interpretability, and how emerging tools are accelerating translational microbiome research.

|
Scooped by
mhryu@live.com
January 11, 11:52 AM
|
Lignin, a complex natural aromatic polymer, poses significant challenges to its efficient degradation, hindering the utilization of biomass for many industrial applications. Bacterial degradation of lignin may offer a promising solution to this challenge. This project aimed at elucidating the function of secreted oxidative enzymes from Pseudomonas putida involved in degradation and utilization of lignin and lignin-derived compounds. Using CRISPR-Cas9 and CRISPR-Cas3 systems, the putative lignin-degrading versatile peroxidase gene (VP; PP_1686, originally annotated as glutathione peroxidase GPx) and dye-decolorizing peroxidase gene (PP_3248) were individually knocked out from P. putida KT2440. The ∆PP_1686 mutant exhibited impaired growth and utilization of lignin-derived compounds. This correlated with reduced expression of p-hydroxybenzoate hydroxylase pobA and of DNA repair modules, alongside compensatory upregulation of energy and redox supply pathways. This work expands our knowledge on bacterial glutathione peroxidase by presenting a role beyond ROS scavenging. This work revealed the importance of P. putida VP/GPx in maintaining redox balance while supporting lignin-derived aromatic metabolism, offering new targets for future investigation into stress–metabolism crosstalk and lignin valorization strategies.

|
Scooped by
mhryu@live.com
January 11, 11:44 AM
|
RNA detection applications can be augmented if a sensed RNA can be directly functionally transduced. However, there is no generalizable approach that allows an RNA trigger itself to directly activate diverse non-coding RNA effectors. Here, we report engineering of a programmable, RNA trigger-activated, dual-site self-cleaving ribozyme with modular sensing domain and cleavage product. This platform, UNlocked by Activating RNA (UNBAR), is entirely encoded within one RNA strand. The ribozyme can be designed to be almost completely inactive in absence of trigger, and to exhibit single-nucleotide trigger specificity. UNBAR ribozymes carry out cell-free sensing and protein-free amplification of microRNA and viral RNA sequences, and trigger-dependent release of ncRNA effectors sgRNA, shRNA and aptamer. We demonstrate RNA detection and functional transduction by a cleaved aptamer, whose fluorescence can be directly read out as a function of trigger RNA. We further engineer the ribozyme for function in cells, and demonstrate trigger-dependent regulation of CRISPR-Cas9 editing by sgRNA-embedded ribozymes in zebrafish embryos and human cells. UNBAR is a first-in-class modality with potential to be developed into a versatile platform for synthetic biology, diagnostics and gene regulation. To enable a sensed RNA to activate diverse RNA effectors, the authors engineer a programmable dual-site ribozyme that, upon RNA trigger binding, self-cleaves to release an embedded RNA. It enables trigger-dependent release of diverse ncRNAs and controls CRISPR-Cas9 editing in zebrafish and human cells.

|
Scooped by
mhryu@live.com
January 10, 4:46 PM
|
The pathogenic bacterium Vibrio parahaemolyticus represents a substantial economic and public health concern; however, elucidating its virulence mechanisms has been significantly impeded by its inherent resistant to genetic manipulation, primarily attributed to sophisticated immune defense systems including restriction-modification (R-M) modules, CRISPR-Cas systems, standalone DNases, and DdmDE systems. Paradoxically, while genetic modification is essential for overcoming these barriers, the very barriers themselves obstruct DNA introduction. Our investigation focused on the V. parahaemolyticus X1 strain, where initial plasmid transformation attempts proved unsuccessful. However, low-efficiency conjugation allowed knockout of defense genes, thereby silencing the host’s defense mechanisms. Our findings revealed a standalone DNase, Vpn, as the predominant obstacle to foreign DNA entry in the X1 strain, while a DdmDE system executes elimination of invaded plasmids. Leveraging these insights, we created the V. parahaemolyticus X2 strain via sequential depletion of the Vpn nuclease and the DdmDE system. Capitalizing on the bacterium’s exceptional growth rate, characterized by a generation time of approximately 10.5 min, we established a highly efficient molecular cloning platform capable of creating a new plasmid construct within a single day. This work not only presents a strategic framework for genetic manipulation of previously recalcitrant bacterial species but also underscores the potential of fast-growing marine bacteria as promising candidates for next-generation biotechnological applications.
|
 Your new post is loading...
Your new post is loading...