Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 3:40 PM
|

|
Scooped by
mhryu@live.com
Today, 3:30 PM
|
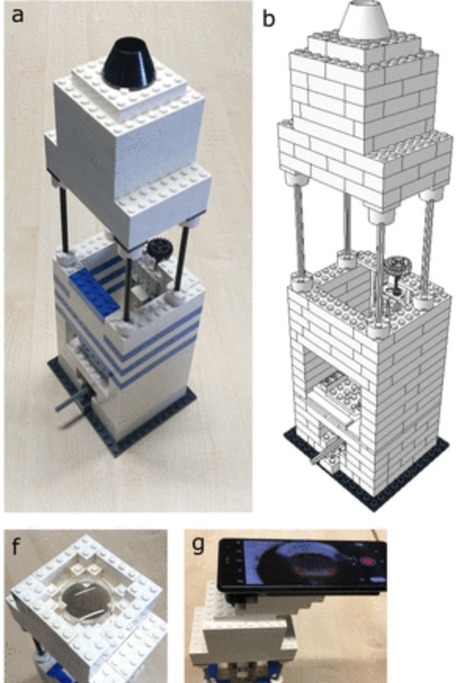
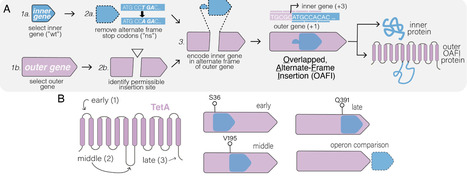
Overlapping genes—wherein two different proteins are translated from alternative reading frames of the same DNA sequence—provide a means to stabilize an engineered gene by directly linking its evolutionary fate with that of an overlapping gene. However, creating overlapping gene pairs is challenging, as it requires redesigning both protein products to accommodate overlap constraints. Here, we present a new “overlapping, alternate-frame insertion” (OAFI) method for creating synthetic overlapping genes by inserting an “inner” gene, encoded in an alternate frame, into a flexible region of an “outer” gene. Using OAFI, we create new overlapping gene pairs of genetic reporters and bacterial toxins within an antibiotic resistance gene. We show that both the inner and outer genes retain function despite redesign, with translation of the inner gene influenced by its overlap position in the outer gene. Importantly, we show that, despite these inner gene sequences not contributing to outer gene function, selection for the outer gene alters the permitted inactivating mutations in the inner gene, and that overlapping toxins can restrict horizontal gene transfer of the antibiotic resistance gene. Overall, OAFI offers a versatile tool for synthetic biology, expanding the applications of overlapping genes in gene stabilization and biocontainment.

|
Scooped by
mhryu@live.com
Today, 2:52 PM
|
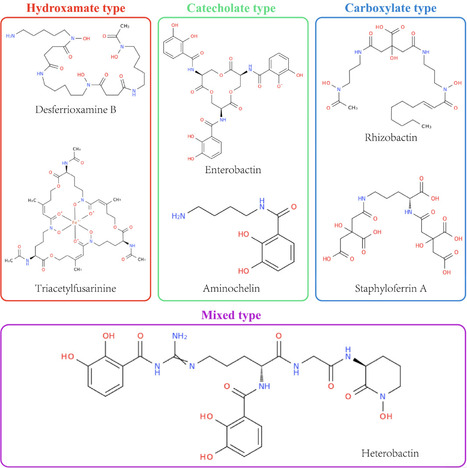
Iron is an essential micronutrient for nearly all microorganisms, yet its bioavailability is severely limited in most environments. To overcome this restriction, microorganisms produce siderophores, high-affinity iron-chelating molecules that play pivotal roles in microbial iron homeostasis, interspecies competition, and host–pathogen interactions. Despite extensive research, current understanding of siderophore biosynthetic and regulatory diversity remains largely limited to specific models, with comprehensive cross-taxonomic frameworks only beginning to emerge. This review systematically integrates recent advances in the genetic and biochemical foundations of microbial siderophore production, focusing on the two major biosynthetic pathways: nonribosomal peptide synthetase (NRPS)-dependent and NRPS-independent synthetase (NIS). We further elaborate on the diverse transport systems in Gram-negative and Gram-positive bacteria, as well as fungi, alongside the iron-responsive regulators (e.g., Fur) and gene clusters that coordinate iron uptake and utilization. Beyond physiological mechanisms, we discuss how these insights inform emerging applications of siderophores across multiple fields: in medicine, enabling “Trojan horse” antimicrobial strategies; in agriculture, enhancing plant iron uptake and serving as biocontrol agents; in environmental remediation, facilitating heavy-metal detoxification; and in biosensing, acting as selective probes for metals and pathogens. By bridging fundamental mechanisms with practical applications, this review aims to provide an integrative perspective for future exploration of microbial iron homeostasis and its biotechnological potential.

|
Scooped by
mhryu@live.com
Today, 2:30 PM
|
The growing global demand for food is limited by low seed germination rates, a key constraint in crop production. Priestia megaterium W101, a plant growth-promoting rhizobacterium with high indole-3-acetic acid (IAA) yield, was found to significantly promote the germination of wheat seeds and the growth of the root system. For the first time, we characterized in Priestia megaterium a complete indole-3-pyruvic acid (IPyA) pathway, encoded by ipdC, patB, feaB, and gene1566, together with a yedL-encoded alternative pathway, through integrated multiomics analyses and CRISPR/Cas9-mediated heterologous expression. In addition, we reconstructed the complete IPyA pathway in Bacillus subtilis 168, which increased IAA production by approximately 98% compared to that in the wild-type strain. Overall, this study elucidates the IAA biosynthetic network in W101 and highlights its potential as a core strain for sustainable microbial inoculant development in green agriculture.

|
Scooped by
mhryu@live.com
Today, 1:53 PM
|
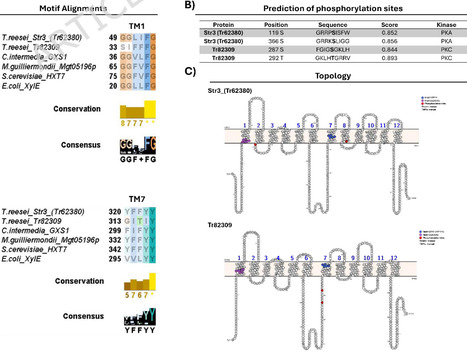
The engineering of Saccharomyces cerevisiae for the use of xylose is fundamental to improving fermentation performance in the production of second-generation ethanol (2G) via pentose fermentation. For this, one of the main strategies involves expressing heterologous xylose transporters to ensure efficient uptake of this sugar. However, due to the intrinsic non-specificity of sugar transporters, competition occurs between sugars (e.g., xylose and glucose), leading to reduced pentose transport efficiency and lower ethanol productivity. This study aimed to develop and characterize sugar transporters with lower affinity for glucose, while maintaining the ability to transport xylose, through genetic improvement of Trichoderma reesei transporters for heterologous expression in S. cerevisiae. To this end, alignments were made to find motifs described as important for xylose transport, and phosphorylation sites were predicted to achieve the objective. Based on these predictions, the transporters were modeled and docked with glucose and xylose. The transporters with the desired phenotype were expressed in S. cerevisiae strains for characterization. Drop assays and aerobic fermentation trials were performed to confirm the predicted profile. In silico analysis shows that two mutations in Str3 (Tr62380) exhibited a promising phenotype. For Tr82309, which has not yet been characterized, it was decided to proceed with characterizing the wild transporter. Drop assays revealed qualitative differences in growth phenotypes among the tested transporters. Docking was used strictly as a qualitative, hypothesis-generating tool to prioritize mutations for experimental testing and was not intended to predict transport kinetics or substrate affinity. The mutants of Str3 (Tr62380) did indeed lose their natural affinity for hexoses. Additionally, Tr82309 showed limited activity in liquid assays but supported growth at higher xylose concentrations in solid medium, suggesting condition-dependent functionality rather than confirmed substrate specificity. In the aerobic fermentation assay, only Str3 (Tr62380)_WT exhibited high efficiency in the uptake of sugars from the medium; mutations inserted in Str3 (Tr62380) reduced the ability to transport sugars, primarily glucose. Phosphorylation mimetics provided in vivo evidence that post-translational modification can influence the substrate utilization profile of sugar transporters. Docking served as a qualitative screening step to guide the selection of mutations for experimental validation. We also present phosphorylation sites as a new target for engineering studies of sugar transporters. However, experimental validation is indispensable. In summary, our findings provide a rational framework for the engineering of glucose-insensitive xylose transporters, enabling more efficient co-fermentation in biorefineries.

|
Scooped by
mhryu@live.com
Today, 1:25 PM
|
How long do fungal hyphae persist in the environment? And how does this differ between groups and species of fungi? Despite growing knowledge of fungal contributions to decomposition and soil carbon cycles, surprisingly little is known about the turnover of mycelia: What happens to fungal hyphae over time? And how this impacts different fungi's contribution to carbon sequestration? In this study, we compared microscale persistence of fungal hyphae using microfluidic chip technology and visual quantification of hyphal degradation and turnover across six different wood-decay Basidiomycete species. Measured traits included hyphal extension, coverage, turnover rates, and changes in hyphal morphology over time when supplied with two carbon sources of differing recalcitrance. Species clustered into two groups: one with a frugal nutrient strategy (high turnover capacity, active persistence of cytoplasmic hyphae) and one with a wasteful strategy (low turnover of hyphae and large remnants of skeletonized hyphae). Differences matched the ephemeral or long-lasting nature of their fruiting bodies and the substrates they inhabit. Carbon type also influenced hyphal persistence over time. Our results suggest that hyphal turnover has a genetic basis linked to species ecology yet is also shaped by environmental factors such as carbon availability, highlighting the dynamic nature of fungal mycelia.

|
Scooped by
mhryu@live.com
Today, 1:16 PM
|
Plant-biotic interactions are driven by the exchange of molecules. Small peptide hormones like CLAVATA3/EMBRYO SURROUNDING REGION (CLE) peptides play central regulatory roles in these interactions. CLEs determine the extent of symbiotic interaction to balance costs and benefits for the host. In parasitic interactions, CLEs regulate the formation of feeding sites by plant pathogenic nematodes and promote the formation of haustoria in parasitic plants. By reviewing recent findings on CLE functions, their receptors, and responses across different biotic interactions, we provide insights into the increasingly complex roles of CLEs in plant development and nutrient signaling.

|
Scooped by
mhryu@live.com
Today, 9:59 AM
|
Recombinant E. coli with nitrogen-fixation activity has been constructed using a simple approach of introducing a minimal gene set of nitrogen fixation genes along with its upstream region from Paenibacillus species; however, limitations may exist in nif expression. To understand the limitations of heterologous mass production of nitrogenase in E. coli, we evaluated this approach using various nif operons cloned from Paenibacillus species. Initial attempts revealed that, despite the high nitrogenase activity observed in the parental strains, the corresponding nif operons did not consistently function in E. coli. Further analysis revealed that certain upstream regions of the nif operons function as constitutive σ70-dependent promoters in E. coli, and the host acquired nitrogenase activity only when the upstream region acted as a constitutive promoter. Notably, recombinant E. coli achieved high nitrogenase activity by linking a functional upstream region to the nif operon from a highly active parental strain. Additional improvements in nitrogenase activity were achieved by combining promoter replacement with synthetic biology approaches to enhance electron transfer from glucose metabolism to nitrogenase and metal cluster assembly for protein maturation. These findings provide new insights into heterologous nitrogenase production in E. coli for sustainable nitrogen fixation.

|
Scooped by
mhryu@live.com
February 5, 11:45 PM
|
We used an autonomous lab, comprising a large language model (LLM) and a fully automated cloud laboratory, to optimize the cost efficiency of cell-free protein synthesis (CFPS). By conducting iterative optimization, the LLM-driven autonomous lab was able to achieve a 40% reduction in the specific cost ($/g protein) of CFPS relative to the state of the art (SOTA). This cost reduction was accompanied by a 27% increase in protein production titer (g/L). Iterative experimental design, experiment execution, data capture and analysis, data interpretation, and new hypothesis generation were all handled by the LLM-driven autonomous lab. The interface between OpenAI's GPT-5 LLM and Ginkgo Bioworks' cloud laboratory incorporated built-in validation checks via a Pydantic schema to ensure that AI-designed experiments were properly specified. Experimental designs were translated into programmatic specification of multi-instrument biological workflows by Ginkgo's Catalyst software and executed on Ginkgo's Reconfigurable Automation Cart (RAC) laboratory automation platform, with human intervention largely limited to reagent and consumables preparation, loading and unloading. By integrating LLMs with programmatic control of a cloud lab, we demonstrate that an LLM-driven autonomous lab can successfully perform a real-world scientific task, highlighting the potential of AI-driven autonomous labs for scientific advancement.

|
Scooped by
mhryu@live.com
February 5, 11:24 PM
|
Biomass separation represents a critical bottleneck in Komagataella phaffii-based biopharmaceutical processes, as typically high cell densities of 40 - 50 % create significant operational, technical and economic challenges for harvest operations. Yeast cell aggregation (flocculation) provides a solution to accelerate cell sedimentation by increasing particle size, thus allowing to improve biomass-supernatant separation efficiency during both natural gravity settling and (continuous) centrifugation operations. This study demonstrates successful engineering of K. phaffii strains with an inducible flocculation phenotype using CRISPR/Cas9-based genome editing to integrate the Saccharomyces cerevisiae FLO1 (FLO1) gene under control of various regulatory elements, including methanol-inducible and derepressible promoters. Flocculation strength could be enhanced by implementing transcriptional positive feedback circuits based on the methanol-inducible AOX1 promoter. To address methanol-free production requirements, we developed alternative systems to retrofit PAOX1-based ScFLO1 expression and exploited the derepressible PDF promoter, offering broader compatibility with biopharmaceutical manufacturing facilities. Flocculating cells cultivated in a bioreactor demonstrated significantly improved sedimentation behavior, with considerably lower supernatant turbidity after short low-speed centrifugation compared to non-flocculating controls. Crucially, cell flocculation had no negative impact on product amount and quality when expressing a multivalent NANOBODY® VHH molecule with pharmaceutical relevance. Thus, this work establishes the first genetically engineered flocculation system in K. phaffii compatible with recombinant protein production, providing the basis for an innovative approach to streamline harvest operations in biopharmaceutical processes.

|
Scooped by
mhryu@live.com
February 5, 11:14 PM
|
In synthetic biology, DNA assembly is a routine process where increasing demands for standardization, high-throughput capacity, and error-free execution are driving the development of accessible, automated solutions. Here, we present Slowpoke, a user-friendly and flexible workflow for Golden Gate-based cloning designed for the popular entry-cost, open-source liquid-handling platforms Opentrons OT-2 and Flex. Slowpoke automates the key steps of the DNA assembly process, including cloning, E. coli transformation, plating, and colony PCR, requiring user intervention primarily for colony picking and plate transfers. To further simplify the usage, we developed a free graphical user interface (GUI), available at https://slowpoke.streamlit.app/, which enables rapid protocol generation through simple file uploads. We validated the workflow using two Golden Gate-based toolkits, the MoClo Yeast Toolkit (YTK), and SubtiToolKit (STK). High assembly efficiencies were achieved across platforms for basic transcript unit constructions: 17/17 positive colonies with YTK on OT-2, 11/12 on Flex, and 8/13 with STK on OT-2. High-throughput assemblies were also performed with six parts in Flex using YTK-compatible parts, and 55 out of 57 combinations resulted in correct constructs. These results confirm the robustness and adaptability of the workflow across toolkit complexity and automation platforms. The Slowpoke suite, including code scripts and templates, is freely available at https://github.com/Tom-Ellis-Lab/Slowpoke, offering an accessible and modular solution for automating Golden Gate cloning in synthetic biology laboratories.

|
Scooped by
mhryu@live.com
February 5, 11:06 PM
|
The expansion of sequencing technologies and bioinformatics has greatly advanced our understanding of microbial “dark matter,” yet the isolation of pure cultures, especially among Archaea, remains rare and challenging. Cultivation is still essential for the reliable characterization of microbial metabolism, which cannot be fully replaced by metagenomics and other omics-based approaches. Here, we report the first cultivated representatives of a deep-branching archaeal lineage previously known as Candidatus Marsarchaeota. Our phylogenomic analyses place these isolates within the phylum Thermoproteota as a novel order, Tardisphaerales. Members of Tardisphaerales dominate the prokaryotic communities in acidic hot springs below 70°C, comprising up to 40% of the total microbial population, underscoring their ecological significance. Functional genomics and culture experiments reveal a thermoacidophilic, anaerobic lifestyle, with energy metabolism based on carbohydrate fermentation, particularly of polysaccharides. This metabolic capability is supported by numerous glycosidase-encoding genes and by unprecedented metabolic versatility among thermoacidophiles. The isolates possess complete glycolysis, Entner-Doudoroff, and pentose-phosphate pathways, allowing them to utilize different sugars. Specialization in polysaccharide hydrolysis presumably provides an adaptive advantage for these slow-growing archaea, as most other heterotrophic thermoacidophiles prefer peptides or simple sugars. Furthermore, robust defense mechanisms against reactive oxygen species and persistence in acidic conditions enable Tardisphaerales to outcompete other heterotrophs and maintain dominance in these extreme habitats. The discovery and cultivation of this new order expand prokaryotic taxonomy and reveal the key players in carbon cycling in acidic geothermal ecosystems.

|
Scooped by
mhryu@live.com
February 5, 8:56 PM
|
Photosymbioses provide carbon and oxygen to the biosphere, yet the mechanisms underlying their evolution remain poorly understood. We develop a naive system based on the predatory ciliate Tetrahymena thermophila, not known for hosting symbionts, to recapitulate early events of photosymbiosis evolution. T. thermophila readily phagocytoses eukaryotic algae (Chlorella variabilis) or cyanobacteria (Synechococcus elongatus). Feeding on either prey in a low-carbon medium provided little or no growth advantage. By contrast, in a hypoxic environment, both intracellular C. variabilis and S. elongatus can support temporary survival of T. thermophila. These results suggest that oxygen supply within the host could represent a more plausible initial advantage supporting photosymbiosis evolution than carbon metabolites. While most extant photosymbioses are based on carbon supply to the host cell, we therefore propose that this would be a secondary event occurring from initial evolution in anoxic or hypoxic conditions, where O2 production is crucial for establishing the initial steps of photosymbiosis.
|

|
Scooped by
mhryu@live.com
Today, 3:34 PM
|
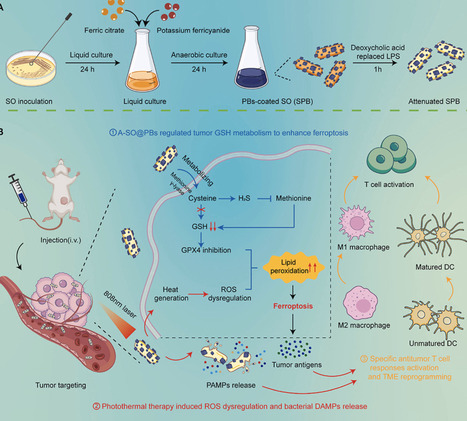
Bacterial combination therapy offers immense promise for treating aggressive "cold" solid tumors, such as triple-negative breast cancer (TNBC). However, the clinical translatability of traditional genetic engineering is often hampered by operational complexity and genetic instability. Here, we developed A-SPB—a non-genetically engineered, multi-functional living bioreactor based on Shewanella oneidensis MR-1 (SO), surface-modified with Prussian blue (PB) nanoparticles and attenuated via deoxycholic acid (DA) treatment. This platform serves as an in situ metabolic factory that "hijacks" the sulfur-metabolism within the hypoxic tumor microenvironment. By actively depleting cysteine, A-SPB not only starves the glutathione (GSH) synthesis pathway but also generates hydrogen sulfide (H2S) to inhibit the transsulfuration bypass, leading to a dual-pronged collapse of the GSH/GPX4 antioxidant axis and triggering robust tumor ferroptosis. This metabolic sensitization is further amplified by the PB-mediated photothermal therapy (PTT), which generates localized hyperthermia and excessive reactive oxygen species (ROS). Notably, the PTT serves as a dual-functional "bio-switch": it promotes acute tumor ablation while simultaneously triggering bacterial self-lysis to ensure biosafety. This programmed lysis releases a synergistic cocktail of bacterial PAMPs and tumor-derived DAMPs, which effectively remodels the immunosuppressive TME and initiates a potent systemic antitumor immune response. By integrating metabolic reprogramming, sensitized ferroptosis, and on-demand immune activation through simple surface engineering, this study provides a highly translatable and safe paradigm for the next generation of living bacterial therapeutics.

|
Scooped by
mhryu@live.com
Today, 3:01 PM
|
Microbes inhabiting soils experience periodic water deprivation. The effects of desiccation on DNA, protein, and membrane integrity are well-described. However, the effects of drying and rehydration on the composition of cellular RNA and metabolites are still poorly understood. Here, we describe how slow drying and rehydration with water vapor influence the composition of RNAs and metabolites in a soil Arthrobacter. While drying reduced cultivability relative to hydrated controls, water vapor rehydration fully restored it. Ribosomal RNA proportions remained constant throughout all treatments, and mRNA profiles showed stable composition during desiccation—changing only during transitions into and out of desiccation-induced dormancy. Six transcriptional modules displayed distinct expression patterns in desiccated-rehydrated samples relative to hydrated controls, including desiccation-rehydration responsive and rehydration-specific profiles. Targeted intracellular metabolomics revealed similarly static profiles during desiccation, with a cluster of ribonucleosides and nucleobases increasing in response to desiccation and returning to baseline levels upon rehydration with water vapor. These findings demonstrate that both mRNA and metabolite profiles remain essentially frozen in desiccated Arthrobacter, with dynamic changes occurring only during state transitions. These results have important implications for environments with frequent drying cycles where stable mRNA in dormant cells combined with intracellular RNA recycling may obscure interpretations of RNA-based environmental analyses that use RNA as a marker of microbial activity. Our results suggest that RNA-based activity assessments in periodically dry environments require careful consideration of dormancy-associated molecular preservation.

|
Scooped by
mhryu@live.com
Today, 2:30 PM
|
Cassava production in sub-Saharan Africa is severely impacted by diseases. Most pathogens require interaction with host susceptibility factors to complete their life cycles and cause disease. Targeted DNA methylation is an epigenetic strategy to alter gene expression in plants, and we previously reported that a zinc-finger fused to DMS3 could establish methylation at the promoter of MeSWEET10a, a bacterial susceptibility gene, and this resulted in decreased disease. Here, we attempt a similar strategy for cassava brown streak disease. This disease is caused by the ipomoviruses CBSV and UCBSV. These viruses belong to the family Potyviridae, which has been shown extensively to require host eIF4E-family proteins to infect plants and cause disease. We previously found that cassava plants with simultaneous knockout mutations in two eIF4E genes, nCBP-1 and nCBP-2, resulted in decreased susceptibility to CBSD. Here, we report successful simultaneous targeting of both promoters with methylation using a dCas9-DRMcd-SunTag system. However, in contrast to our previous work with MeSWEET10a, controls indicate that CRISPR interference is occurring in these lines and is sufficient for the reduction of gene expression. Future research will use genetic crosses to segregate away the DNA methylation reagents and, if DNA methylation proves heritable, assess whether methylation alone is sufficient to increase resistance to CBSD.

|
Scooped by
mhryu@live.com
Today, 2:00 PM
|
This work constructed a recombinant lactic acid bacterium secreting β-galactosidase for GOS formation during milk fermentation. First, GalINF, a β-galactosidase derived from infant feces, was characterized to effectively produce GOS in milk, reaching a content of 10.03 g/L. To export GalINF in Lactococcus lactis, six signal peptide candidates were employed, resulting in extracellular activities as low as 52.83–85.65 U/L. Then, GalINF (114.6 kDa) was split into two complementary modules, M1-P723 and A724-I1023, which could be independently secreted and actively reconstituted with the help of the protein scaffold SpyCatcher/SpyTag. The resultant Lc. lactis B1RG exhibited an extracellular β-galactosidase activity of 544.22 U/L. Fermentation of pasteurized milk with Lc. lactis B1RG and the traditional yogurt starters reduced lactose to 19.67 g/L and yielded 7.17 g/L GOS. This work established an effective strategy to export large-sized proteins extracellularly and demonstrated the applicability of LAB secreting β-galactosidase for GOS-enriched fermented dairy products.

|
Scooped by
mhryu@live.com
Today, 1:35 PM
|
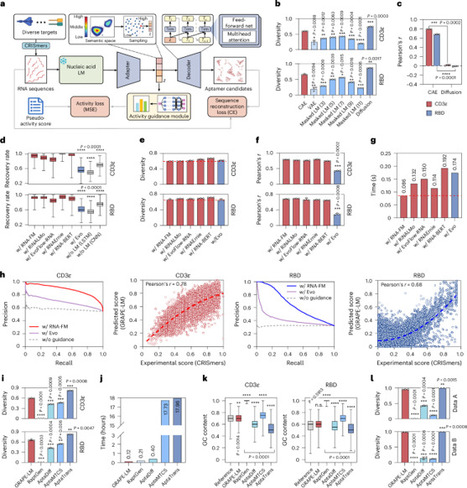
The directed evolution of biomolecules is an iterative process. Although advancements in language models have expedited protein evolution, effectively evolving RNA remains a challenge. RNA aptamers, selected for their binding properties, provide an ideal system to address this challenge, yet traditional aptamer discovery still relies on labor-intensive, multi-round screening. Here we introduce GRAPE-LM (generator of RNA aptamers powered by activity-guided evolution and language model), a generative artificial intelligence framework designed for the one-round evolution of RNA aptamers. GRAPE-LM integrates a transformer-based conditional autoencoder with nucleic acid language models and is guided by CRISPR−Cas-based aptamer screening data derived from intracellular environments. We validate GRAPE-LM on three disparate targets: the human T cell receptor CD3ε, the receptor-binding domain of the SARS-CoV-2 spike protein and the human oncogenic transcription factor c-Myc (an intracellular disordered protein). GRAPE-LM, informed with only a single round of CRISPR−Cas-based screening, successfully obtains RNA aptamers that outperform those driven from multiple rounds of human selection and optimization. Combining generative AI and one round of wet lab evolution generates high-affinity RNA aptamers.

|
Scooped by
mhryu@live.com
Today, 1:21 PM
|
Crop protection against soilborne pathogens such as root-knot nematodes (Meloidogyne spp.) has been a losing battle on too many occasions. Often going undetected until they cause major crop losses, these organisms have been conventionally managed by field application of synthetic nematicides, with high environmental cost. Agroecologically sound practices for nematode control, including soil organic matter management, crop diversification and intercropping, are being developed and tested, but the underlying mechanisms behind successful control have remained elusive. In an article, Hama et al. (2025; doi:10.1111/nph.70789) have uncovered a form of plant resistance that may be responsible for the complex and often unanticipated effects in the field: plants can transfer defence compounds between them, causing significant metabolome changes and, ultimately, inducing root-knot nematode resistance in otherwise susceptible neighboring plants. benzoxazinoids (BXs) exuded by rye (Secale cereale) roots into the soil can be taken up, metabolised and translocated by roots of clover (Trifolium repens)

|
Scooped by
mhryu@live.com
Today, 1:09 PM
|
The extent to which microbial processes control soil organic carbon (SOC) dynamics remains uncertain. Carbon use efficiency (CUE), that is, the fraction of assimilated carbon allocated to growth, has been used as a key parameter but its relationship with SOC reflects carbon partitioning rather than the absolute magnitude of microbial fluxes. The microbial growth rate could provide a more mechanistic link to SOC accumulation because it quantifies biomass production and reflects necromass formation. Here we combine a global ¹⁸O–H2O dataset (n = 268 paired observations) with outputs from four land surface models to test whether growth rate predicts SOC more strongly than CUE. In the incubation experiments, growth rates are more closely associated with SOC than CUE, although soil properties and climate explain equal or greater variance. Models reproduce the stronger role of growth rate over CUE but tend to underestimate the abiotic controls. The models also emphasize CUE as the main predictor of the SOC-to-net primary production ratio, in contrast to observations, which indicates the soil’s capacity to retain plant carbon inputs. Together, these findings identify the microbial growth rate as a diagnostic that can help bridge models with empirical data and guide a more balanced representation of microbial and mineral controls in SOC projections. Microbial carbon use efficiency is a strong predictor of soil organic carbon stocks. Here the authors reveal that the microbial growth rate is a more reliable and informative predictor, and that modelling approaches tend to overemphasize the role of biotic over abiotic controls compared to empirical data.

|
Scooped by
mhryu@live.com
Today, 9:33 AM
|
Taxonomic classification alone fails to capture the ecological and functional diversity of vaginal microbiomes, particularly those dominated by Gardnerella species. Using the expanded VIRGO2 gene catalog, we developed the vaginal inference of subspecies and typing algorithm (VISTA), a novel ortholog-based framework that defined metagenomic subspecies and 25 metagenomic community state types (mgCSTs), including six distinct Gardnerella-dominated profiles. The mgCSTs exhibit marked differences in species composition, functional gene content, transcriptional activity, and host immune responses. These findings reveal that Gardnerella predominance does not uniformly equate to dysbiosis and underscore the importance of functional context in shaping host-microbiome interactions. VISTA provides scalable classifiers and an interactive application to support mechanistic studies of vaginal microbiome function and its implications for reproductive health.

|
Scooped by
mhryu@live.com
February 5, 11:35 PM
|
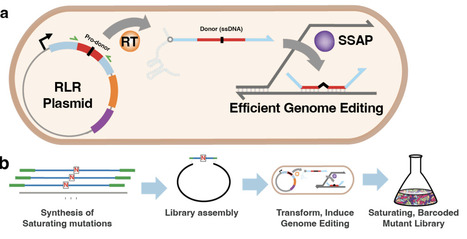
Saturation mutagenesis is a powerful tool for understanding and engineering the function of biological systems, and has been applied successfully to characterize the mutational landscape of individual proteins and genetic loci. However, it has not been applied at the whole-genome scale due to the challenges of both creating and quantifying a saturating set of mutations. Here we introduce Biobloom, a retron-based method for barcoded saturation mutagenesis at the scale of a whole bacterial genome. We constructed a barcoded Biobloom library with >99% projected sampling of saturating single-nucleotide polymorphism (SNP) mutations of the E. coli genome, and applied it to identify beneficial mutations under salt and antibiotic selection. Relative to other techniques like CRISPR-enabled mutagenesis or Adaptive Laboratory Evolution, Biobloom excels at identifying diverse causal SNPs quickly and at smaller working volumes. A barcoded, saturating mutation library is also a shared resource, and we are releasing the updated Biobloom-E.coli-2.0 library to the scientific community for broader adoption and application. Together, Biobloom makes barcoded saturation mutagenesis accessible at whole-genome scale, creating new opportunities for large-scale data collection and bacterial engineering.

|
Scooped by
mhryu@live.com
February 5, 11:20 PM
|
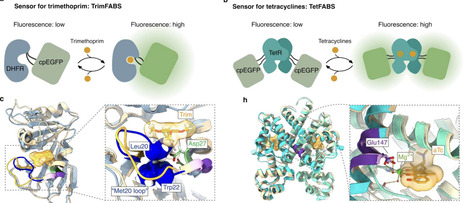
Antibiotic treatment can fail due to insufficient drug availability at the site of infection or limited accumulation within bacterial pathogens. However, it is poorly understood how antibiotics penetrate infected tissues and complex bacterial aggregates, limiting insights into the mechanisms of treatment failure. Here, we present genetically-encoded allosteric biosensors for two antibiotic classes, trimethoprim and tetracycline, which enable real-time monitoring of antibiotic concentrations inside bacterial cells. The biosensors consist of circularly permuted EGFP linked to the sensory domains DHFR or TetR. To extend this approach to low oxygen environments, we engineered an oxygen-independent trimethoprim biosensor by fusing DHFR to a circularly permuted version of the fluorogenic protein FAST. Using these biosensors, we monitored the antibiotic exposure dynamics of intracellular Salmonella enterica during macrophage infection at the single-cell level, and antibiotic penetration into anaerobic regions of Vibrio cholerae biofilms, as well as antibiotic availability in microoxic conditions in a human bladder tissue model infected with uropathogenic Escherichia coli. These fluorescent biosensors have the potential to be broadly applied for determining antibiotic distributions at infection sites with high spatial and temporal resolution.

|
Scooped by
mhryu@live.com
February 5, 11:10 PM
|
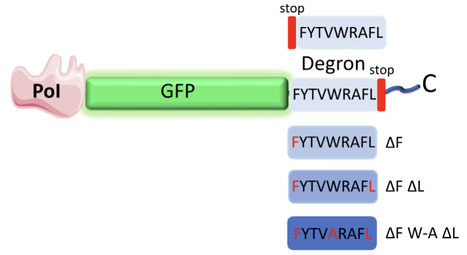
Targeted protein degradation relies on short sequence elements, termed degrons, that direct proteins to the cellular degradation machinery. Here, we report a nine-amino-acid degron (FYTVWRAFL) derived from the 3′ untranslated region (UTR) of human MTCH2 and demonstrate its utility as a tuneable protein-degradation tool. When appended to the C-terminus of proteins, FYTVWRAFL induces rapid and robust degradation by the ubiquitin-proteasome system. Using pharmacological inhibition and genetic perturbation, we show that degradation mediated by this degron is dependent on the E3 ubiquitin ligase MDM2. Structural modeling revealed that FYTVWRAFL interacts with the hydrophobic cleft of MDM2 in a manner similar to the p53 degron. Three hydrophobic residues form the core interaction interface. By systematic mutagenesis of these residues, we generated a panel of degron variants that confer graded levels of protein stability. We demonstrate the versatility of this system by achieving tunable expression of the endogenous protein Elm1 in Saccharomyces cerevisiae. Collectively, our study establishes a compact, transportable, and tunable degron system as a robust toolkit for quantitative control of protein abundance across eukaryotic systems.

|
Scooped by
mhryu@live.com
February 5, 9:00 PM
|
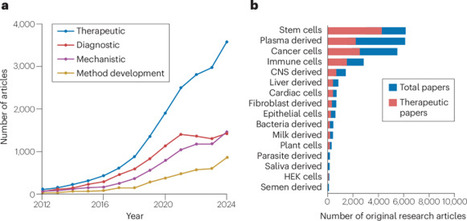
Owing to their natural origin and biocompatibility, extracellular vesicles (EVs) are being recognized as next-generation vehicles for targeted drug delivery. Despite their potential as therapeutic carriers, EVs suffer from heterogeneity, low yields, limited cargo loading efficiency and rapid clearance by the mononuclear phagocyte system. Since the first EV-based clinical trial in 2005, more than 100 clinical trials have investigated the use of EVs as therapeutics and drug carriers. Despite this, no EV-based therapies have received regulatory approval to date. This gap between preclinical research activity and clinical translation underscores persistent scientific challenges and regulatory hurdles that continue to impede the advancement of EV-based therapeutics. In this Review, we examine the research articles published in the field between 2012 and 2024 (38,177 articles), highlighting key developments, persistent challenges and evolving assumptions. We review the current EV landscape and clinical trials, focusing on their organotropism and use as carriers for therapeutics. We compare their advantages and limitations in relation to other nanoparticles, such as lipid nanoparticles and liposomes, and examine how labelling strategies and cell sources influence EV biodistribution. Finally, we outline translational considerations for EV-based therapeutics and propose additional reporting standards, complementing the MISEV 2023 guidelines. Extracellular vesicles (EVs) are emerging as biocompatible carriers for targeted drug delivery, yet challenges in yield, cargo loading and biodistribution persist. In this Review, key trends from over a decade of research are analysed, comparing EVs with lipid-based systems and outlining strategies for improving therapeutic translation and reporting standards.
|
 Your new post is loading...
Your new post is loading...