Your new post is loading...

|
Scooped by
?
Today, 12:06 AM
|
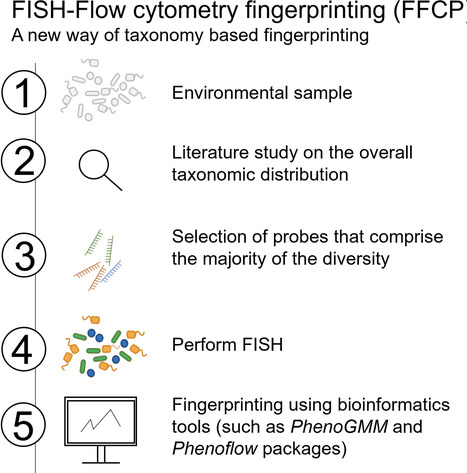
Flow cytometry is a powerful tool to monitor microbial communities, as it allows tracking both changes in the subpopulations and cell numbers at high throughput and a low sample cost. This information can be combined in a phenotypic fingerprint that can be leveraged for diversity analysis. However, as isogenic individuals can manifest phenotypic diversity, for example, due to differing physiological state and phenotypic plasticity, combining the phenotypic information with taxonomic information adds an extra dimension for describing the dynamics of a microbial community. In this research, taxonomic information was incorporated in the microbial fingerprint through fluorescent in situ hybridization (FISH) at a single-cell level. To validate this concept and explore its versatility, two ecosystems with different micro-biodiversity were considered. In the first environment, marine bacteria were monitored for plastic biodegradation in a trickling filter, and in the second, an in vitro simulated human gut microbiome was followed over time. Samples were prepared using different (staining) methods, including FISH, and beta diversity analysis was used to evaluate the level of distinction between differently treated groups in both environments. As a reference to correlate increased distinction with the incorporation of taxonomic information, 16S rRNA gene sequencing was used. Finally, a predictive algorithm was trained to correctly classify samples in the differently treated groups. The results showed that the implementation of FISH in flow cytometry provides more information on a single-cell level to answer specific scientific questions, like distinguishing between phenotypically similar communities or following a specific taxonomic group over time.

|
Scooped by
?
June 23, 11:35 PM
|
Numerous important environments harbor low levels of microbial biomass, including certain human tissues, the atmosphere, plant seeds, treated drinking water, hyper-arid soils and the deep subsurface, with some environments lacking resident microbes altogether. These low microbial biomass environments pose unique challenges for standard DNA-based sequencing approaches, as the inevitability of contamination from external sources becomes a critical concern when working near the limits of detection. Likewise, lower-biomass samples can be disproportionately impacted by cross-contamination and practices suitable for handling higher-biomass samples may produce misleading results when applied to lower microbial biomass samples. This Consensus Statement outlines strategies to reduce contamination and cross-contamination, focusing on marker gene and metagenomic analyses. We also provide minimal standards for reporting contamination information and removal workflows. Considerations must be made at every study stage, from sample collection and handling through data analysis and reporting to reduce and identify contaminants. We urge researchers to adopt these recommendations when designing, implementing and reporting microbiome studies, especially those conducted in low-biomass systems. In this Consensus Statement, the authors outline strategies for processing, analysing and interpreting low-biomass microbiome samples, and provide recommendations to minimize contaminants.

|
Scooped by
?
June 23, 1:10 PM
|
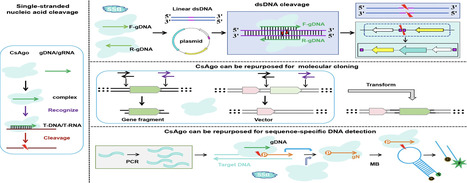
Eukaryotic Argonautes (eAgos) have traditionally been characterized by their ability to utilize RNA guides to identify RNA targets, thereby engaging in post-transcriptional gene silencing pathways. While some eAgos have been demonstrated to use DNA guides for RNA cleavage, the ability of eAgos to cleave DNA targets remains unclear. In this study, we characterized CsAgo, an eAgo protein derived from thermophilic eukaryote Chaetomiumsp. MPI-CAGE-AT-0009, demonstrating a novel ability to cleave both DNA and RNA targets in vitro. Guided by short single-stranded DNA (ssDNA) or RNA, CsAgo exhibits robust RNA cleavage activity at 20–90°C in vitro. CsAgo can effectively cleave ssDNA guided by RNA guides at 20–50°C in vitro. Notably, CsAgo can utilize DNA guides to effectively cleave ssDNA, plasmid double-stranded DNA (dsDNA), and linear dsDNA at ≥80°C in vitro. Based on its ability to cleave dsDNA at high temperatures, CsAgo demonstrates versatility and efficacy in simplifying routine cloning workflows. Additionally, we have developed a CsAgo-based nucleic acid detection method based on a Pyrococcus furiosus Ago-mediated nucleic acid detection method, which exhibits a high sensitivity of six copies/reaction. These results suggest that eAgos include that, in theory, can be utilized for potential DNA-targeting applications. This not only enhances our understanding of eAgos but also expands the toolkit for DNA manipulation.

|
Scooped by
?
June 23, 12:48 PM
|
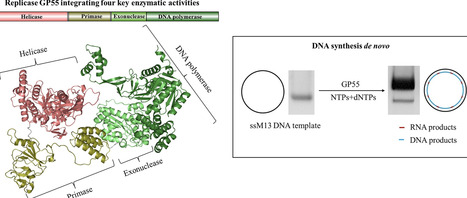
DNA replication is a fundamental process in all living organisms. As the most diverse and abundant biological entities on Earth, bacteriophages may utilize unconventional methods for genome replication. In this study, we identified a novel DNA replicase, GP55, from lactococcal phage 1706. GP55 comprises a helicase domain, a distinctive archaeo-eukaryotic primase domain, and a family B DNA polymerase domain, collectively exhibiting helicase, primase, and DNA polymerase activities, along with intrinsic 3′–5′ exonuclease activity. Notably, the helicase activity of GP55 is UTP/dTTP-dependent rather than ATP-dependent and facilitates strand displacement during DNA synthesis. GP55 exhibits a unique primase activity, recognizing specific but less stringent DNA sequences and preferring GTP for the initiation of RNA primer synthesis. Additionally, a newly identified α-helix domain, composed of two pairs of parallel α-helices, was found to be essential for its primase activity. The multiple activities enable GP55 to efficiently synthesize DNA de novo in the presence of dNTPs and NTPs. This study reveals a concise strategy employed by bacteriophages for genome replication using multifunctional replicases.

|
Scooped by
?
June 23, 11:30 AM
|
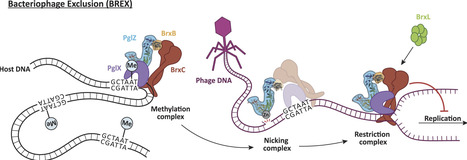
BREX (Bacteriophage Exclusion) systems, identified through shared identity with Pgl (Phage Growth Limitation) systems, are a widespread, highly diverse group of phage defence systems found throughout bacteria and archaea. The varied BREX Types harbor multiple protein subunits (between four and eight) and all encode a conserved putative phosphatase, PglZ, and an equally conserved, putative ATPase, BrxC. Almost all BREX systems also contain a site-specific methyltransferase, PglX. Despite having determined the structure and fundamental biophysical and biochemical behaviours of several BREX factors (including the PglX methyltransferase, the BrxL effector, the BrxA DNA-binding protein, and a commonly-associated transcriptional regulator, BrxR), the mechanism by which BREX impedes phage replication remains largely undetermined. In this study, we identified a stable BREX sub-complex of PglZ:BrxB, generated and validated a structural model of that protein complex, and assessed the biochemical activity of PglZ from BREX, revealing it to be a metal-dependent nuclease. PglZ can cleave cyclic oligonucleotides, linear oligonucleotides, plasmid DNA and both non-modified and modified linear phage genomes. PglZ nuclease activity has no obvious role in BREX-dependent methylation, but does contribute to BREX phage defence. BrxB binding does not impact PglZ nuclease activity. These data contribute to our growing understanding of BREX phage defence.

|
Scooped by
?
June 23, 11:12 AM
|
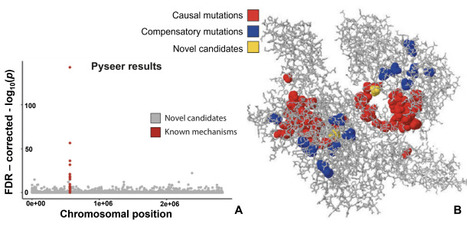
How can we identify causal genetic mechanisms governing bacterial traits? Initial efforts entrusting machine learning models to handle the task of predicting phenotype from genotype yield high accuracy scores. However, attempts to extract meaningful interpretations from the predictive models are found to be corrupted by falsely identified “causal” features. Relying solely on pattern recognition and correlations is unreliable, significantly so in bacterial genomics settings where high-dimensionality and spurious associations are the norm. Though it is not yet clear whether we can overcome this hurdle, significant efforts are being made towards discovering potential high-risk bacterial genetic variants. In view of this, we set up open problems surrounding phenotype prediction from bacterial whole-genome datasets and extending those approaches to learning causal effects, and discuss challenges that impact the reliability of a machine’s decision-making when faced with datasets of this nature. We identify major sources of non-injectivity in the formulation of the genotype-to-phenotype mapping function—linkage-disequilibrium, limited sampling, information loss in representations, unmeasured confounders and observational noise—and analyse their implications for machine learning applications. Using a collection of 4,140 Staphylococcus aureus isolates, we illustrate challenges surrounding the defined open problems.

|
Scooped by
?
June 23, 11:01 AM
|
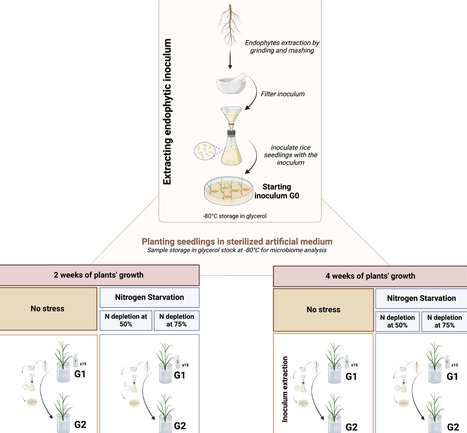
Microorganisms live in close association with plants, forming ecological interaction webs and providing beneficial traits such as nutrition, growth, and tolerance to biotic and abiotic stresses. Via the rhizosphere, plants recruit bacteria which colonize internal plant tissues, creating a spatial gradient between the rhizosphere and the endosphere. This study presents a high throughput in planta endophyte enrichment scheme designed for the identification of 'super'-endophytic bacteria which can serially colonise the rice root endosphere. Oryza sativa (rice) plants were grown in bulk soil, and endophytes were then recovered from roots. The recovered endophyte mixture was used as inoculum for the first generation of rice plantlets, which were then grown under no stress or nitrogen (N) depletion. The total endophytic community was then purified and used as a second inoculum for a new set of plants; this procedure was repeated for four generations. Enrichment patterns of root bacterial endophytes were observed, such as Kosakonia in the non-stressed plants and Ferrovibrio in plants grown under nitrogen starvation. This enrichment method proved to be suitable for the identification of endophytes which can efficiently colonise the root endosphere.

|
Scooped by
?
June 23, 10:50 AM
|
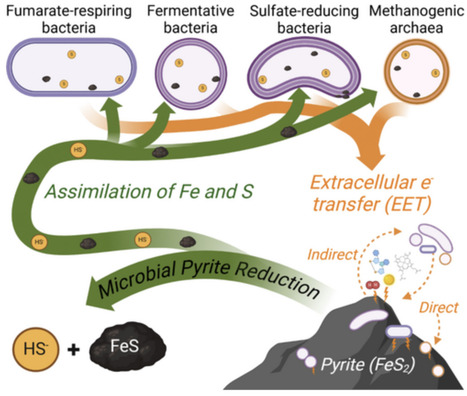
Pyrite, the most abundant iron sulfide mineral in the Earth's crust, has traditionally been considered as a sink for iron and sulfur in the absence of oxygen. Recent research, however, has shown that anaerobic methanogenic archaea can reductively dissolve pyrite and assimilate its products as sources of iron and sulfur. This study explores whether other anaerobic bacteria, including fermentative, nitrate-, iron oxide-, fumarate-, and sulfate-respiring bacteria, can also reduce pyrite and use its dissolution products as sources of iron and sulfur. Results indicate that heterotrophic bacteria respiring fumarate or sulfate, or fermenting organic carbon, can reduce pyrite and assimilate released iron and sulfur. In contrast, nitrate- or iron oxide-respiring cells did not reduce pyrite, suggesting that microbial pyrite reduction is metabolism-specific. All strains capable of reducing pyrite could also use mackinawite as an iron and sulfur source. With the exception of fermentative Bacteroides, strains did not require direct contact with pyrite to reduce the mineral, indicating extracellular electron transfer via electron shuttles. These findings expand the known diversity of microbial groups capable of pyrite reduction and highlight the mineral's lability in various anaerobic environments, with potential implications for the biogeochemical cycles of iron, sulfur, carbon, and oxygen.

|
Scooped by
?
June 23, 10:25 AM
|
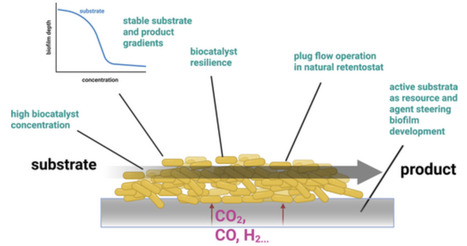
Biofilm-based production systems offer enhanced robustness, higher biomass densities and improved genetic stability compared to traditional stirred tank reactors, presenting promising alternatives for sustainable, efficient biotechnological manufacturing despite challenges in reactor design and process control.

|
Scooped by
?
June 23, 10:18 AM
|
Phenazines are bioactive secondary metabolites with antifungal, anticancer, and insecticidal properties, while hydroxylated derivatives often exhibit enhanced bioactivity. 2-hydroxyphenazine (2-OH-PHZ), which is synthesised by the flavin-dependent monooxygenase PhzO from phenazine-1-carboxylic acid (PCA), shows better bioactivity against the pathogenic fungus Gaeumannomyces graminis vars. tritici. However, the low catalytic efficiency (10%–20% conversion) of PhzO limited 2-OH-PHZ production. To boost PhzO activity, engineering flavin reductase (Fre)-mediated FADH2 regeneration was applied to Pseudomonas chlororaphis LX24AE. Remarkably, this approach improved catalytic efficiency from 25% to 40% and increased the production of a novel dihydroxylated derivative. Then, it was first characterised by UPLC-MS and NMR, and identified as 3,4-dihydroxyphenazine-1-carboxylic acid (3,4-OH-PCA). Next, the Fre-PhzO module through heterologous co-expression in P. putida KT2440 demonstrated a 4.5-fold enhancement in hydroxylation efficiency relative to the PhzO mono-component system, which also confirmed that PhzO and flavin reductase are essential for 3,4-OH-PCA biosynthesis. Moreover, in vitro assays further verified that PhzO exhibits FAD-dependent catalytic promiscuity, simultaneously generating 2-OH-PCA and 3,4-OH-PCA. Furthermore, in vitro and in vivo assays demonstrated that substrate concentration affected the distribution of products. Finally, cytotoxicity evaluation of the isolated 3,4-OH-PCA was performed, and it showed substantial inhibition against oesophageal cancer TE-1 cells and human cervical cancer HeLa cells with an IC50 value of 8.55 μM and 17.69 μM, respectively. This work redefines PhzO as a promiscuous biocatalyst capable of dual hydroxylation, offering a modular platform for engineering bioactive phenazine derivatives.

|
Scooped by
?
June 23, 10:07 AM
|
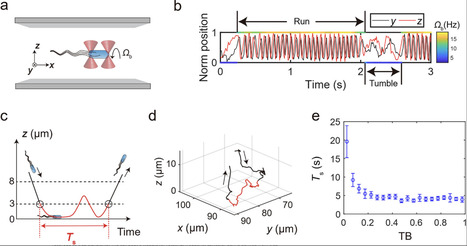
In natural environments, solid surfaces present both opportunities and challenges for bacteria. On one hand, they serve as platforms for biofilm formation, crucial for bacterial colonization and resilience in harsh conditions. On the other hand, surfaces can entrap bacteria for extended periods and force them to swim along circular trajectories, constraining their environmental exploration compared to the freedom they experience in the bulk liquid. Here, through systematic single-cell behavioral measurements, phenomenological modeling, and theoretical analysis, how bacteria strategically navigate these factors is revealed. It is observed that bacterial surface residence time decreases sharply with increasing tumble bias from zero, transitioning to a plateau at the mean tumble bias of wild-type Escherichia coli (≈0.25). Furthermore, it is found that bacterial surface diffusivity peaks near this mean tumble bias. Considering the phenotypic variation in bacterial tumble bias, which is primarily induced by noise in gene expression, this reflects a strategy for bacterial offspring persistence: In the absence of stimulus cues, some bacteria swiftly escape from the nearby surface in case it lacks nutrients, while others, with longer surface residence times, explore this 2D environment most efficiently to find potential livable sites.

|
Scooped by
?
June 23, 9:33 AM
|
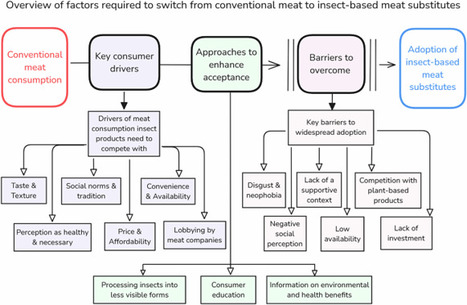
The substantial environmental footprint of meat production means that dietary shifts are needed to reduce greenhouse gas emissions. Insects may offer one alternative, but must first be widely accepted and consumed by the general public. This review evaluates the prospects of insect-based foods to compete with meat. We find that insect-based foods face major challenges, including low consumer acceptance and limited investment. They have a low likelihood of significantly reducing meat consumption, particularly when compared to more accepted plant-based alternatives.

|
Scooped by
?
June 23, 8:45 AM
|
Escherichia coli, a key workhorse in industrial biotechnology, is commonly grown on glucose, which supports rapid growth but leads to acetate overflow, which instead inhibits growth, diverts carbon from the production pathway, and reduces productivity. Recent studies suggest that acetate may also have beneficial effects in glucose-grown E. coli, but its potential in bioprocesses remains unexplored. In this study, we systematically investigated acetate's impact on bioproduction using a kinetic model of glucose and acetate metabolism in E. coli. The model predicts that acetate can enhance bioproduction in glucose-grown E. coli through three mechanisms: (i) by minimizing acetate overflow, thereby reducing carbon loss, (ii) by increasing acetyl-CoA levels, thereby boosting the biosynthetic flux of acetyl-CoA-derived compounds, and (iii) by promoting biomass accumulation, thus improving overall productivity. We experimentally validated the predictions of the model for mevalonate and 3-hydroxypropionate production, where acetate supplementation increased productivity by 117% and 34%, respectively. Our findings provide a valuable framework for optimizing E. coli-based bioprocesses and highlight acetate's underutilized potential in biotechnology. By leveraging acetate from waste streams as a metabolic booster, this approach could contribute to more sustainable and environmentally friendly bioprocesses.
|

|
Scooped by
?
June 23, 11:51 PM
|
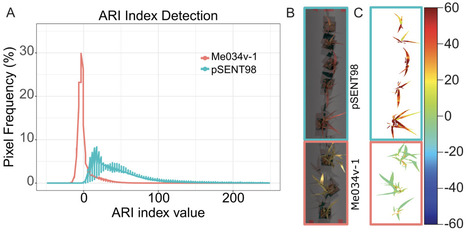
Plant synthetic biology holds great promise for engineering plants to meet future demands. Genetic circuits are being designed, built, and tested in plants to demonstrate proof of concept. However, developing these components in monocots, which the world relies on for grain, lags behind dicot models, such as Arabidopsis thaliana and Nicotiana benthamiana. Here, we show the successful adaptation of a ligand-inducible sensor to activate an endogenous anthocyanin pathway in the C4 monocot model Setaria viridis. We identify two transcription factors sufficient to induce endogenous anthocyanin production in S. viridis protoplasts and whole plants in a constitutive or ligand-inducible manner. We also test multiple ligands to overcome physical barriers to ligand uptake, identifying triamcinolone acetonide (TA) as a highly potent inducer of this system. Using hyperspectral imaging and a discriminative target characterization method in a near-remote configuration, we can non-destructively detect anthocyanin production in leaves in response to ligands. This work demonstrates the use of inducible expression systems in monocots to manipulate endogenous pathways, stimulating plants to overproduce secondary metabolites with value to human health. Applying inducible pigmentation coupled with sensitive detection algorithms could enable crop plants to report on the status of field contamination or detect undesirable chemicals impacting agriculture, ushering in an era of agriculture-based sensor systems.

|
Scooped by
?
June 23, 11:08 PM
|
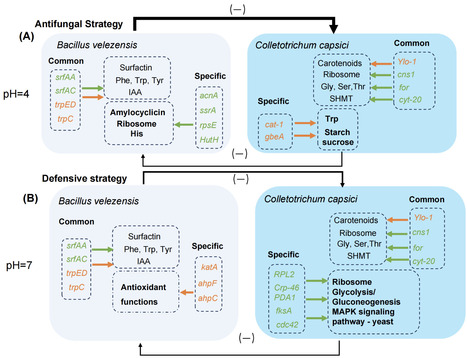
Colletotrichum capsici is the etiological agent of Capsicum anthracnose. Bacillus velezensis has traditionally been recognized as an effective biocontrol agent; however, its efficacy decreases due to soil acidification. In this study, we domesticated Bacillus velezensis XY40-1 along an acid resistance gradient, resulting in a strain capable of growth at pH 4, and might adapt to acidic environments by regulating genes related to spore formation. Notably, the domesticated Bacillus velezensis XY40-1 exhibits significant antagonistic activity against Colletotrichum capsici in acidic dual cultures and effectively reduces the disease index in Capsicum. The domesticated strain employs a direct antifungal strategy under acidic conditions, with the production of amylocyclicin, regulated by acnA, potentially serving as a primary mechanism through which Bacillus velezensis combats Colletotrichum capsici. Conversely, under neutral conditions, domesticated Bacillus velezensis focuses on bolstering its defense mechanisms by increasing the expression of katA, ahpF, and ahpC genes to detoxify peroxides. In addition, a dual RNA-Seq analysis comprehensively investigated the acid tolerance mechanisms and defensive responses of B. velezensis and the pathogenic mechanisms of C. capsici, providing a foundation for the practical application of B. velezensis as a biocontrol agent. These findings offer important insights into the impact of soil acidification on plant disease suppression and contribute to the development of sustainable agricultural practices.

|
Scooped by
?
June 23, 1:02 PM
|
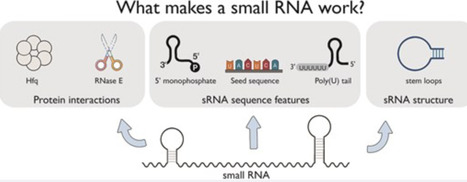
Bacterial small RNAs (sRNA) are key regulators of gene expression, interacting with target messenger RNAs (mRNAs) through imperfect base pairing. Unlike other non-coding RNAs such as microRNAs and PIWI-interacting RNAs, bacterial sRNAs exhibit significant sequence and structural diversity, complicating functional predictions. Recent high-throughput profiling of the sRNA interactome has accentuated this problem by revealing a highly complex network of sRNA interactions. It is clear that there is an incredible diversity of sRNA interactions with different RNA classes in vivo, including different interaction modes with mRNAs. In this review, we attempt to summarize the known sequence and structural features that contribute to sRNA function in bacteria. As many of these features drive recruitment of protein partners, we necessarily focus on interactions with chaperones and ribonucleases, the best studied being Hfq and RNase E. Where possible, we have included examples outside this well-studied system as diversity and rule breaking appear to be central themes of sRNA biology. Understanding the sequences and structures that drive sRNA function will enhance our ability to predict regulatory outcomes, and this may inform the development of effective RNA therapeutics that are inspired by bacterial sRNA mechanisms.

|
Scooped by
?
June 23, 11:36 AM
|
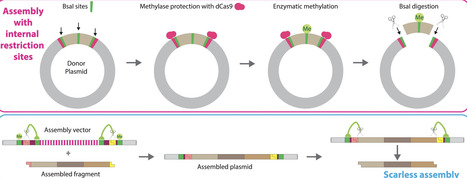
Type IIS restriction enzyme-mediated DNA assembly is efficient but has sequence constraints and can result in unwanted sequence scars. To overcome these drawbacks, we developed UniClo, a type IIS restriction enzyme-mediated method for universal and flexible DNA assembly. This is achieved through a combination of vector engineering, DNA methylation using recombinant methylases, and steric blockade using CRISPR–dCas9 technology to regulate this methylation. Type IIS restriction enzyme sites within fragments to be assembled are methylated using recombinant methylases, while the fragment-flanking outer sites are protected from methylation by a recombinant dCas9–sgRNA complex. During the subsequent assembly reaction, only the protected flanking sites are cut as only they are unmethylated. Fragments are correctly assembled, despite containing internal sites for the single type IIS restriction enzyme used for the one-pot assembly. The assembled plasmid can be used as a donor plasmid in a subsequent assembly round with the same type IIS restriction enzyme and the assembly vector engineering ensures removal of potential scars by a trimming process. This simplifies assembly design and only three vectors are required for any multi-round assembly. These vectors all use the same pair of overhangs. UniClo provides a simple scarless approach for hierarchical assembly of any sequence and has wide potential application.

|
Scooped by
?
June 23, 11:20 AM
|
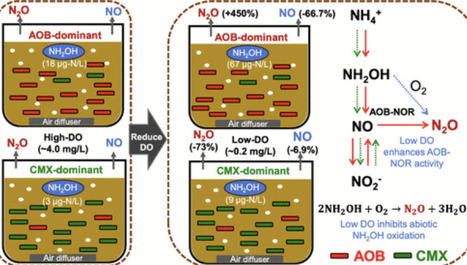
Nitrification significantly contributes to N2O emissions in wastewater treatment, typically enhanced at low dissolved oxygen (DO). The present study revealed that low DO (∼0.2 mg/L) enhanced the N2O emission factor (EF) from nitrification by 4.5 times in canonical ammonia-oxidizing bacteria (AOB)-dominated sludge, while it reduced N2O EF by 73% in comammox Nitrospira-dominated sludge. During nitrification, the accumulation of intermediate NH2OH in AOB-dominant sludge was much higher and increased more significantly by low DO (from 0.018 to 0.067 mg-N/L) compared to comammox Nitrospira-dominant sludge (from 0.004 to 0.009 mg-N/L). In AOB-dominant sludge, the increased NH2OH and upregulated AOB-NOR gene at low DO promoted NO reduction, thus increasing N2O EF while simultaneously decreasing NO emission. In the comammox Nitrospira-dominant sludge, N2O was primarily produced via abiotic pathways. Further investigation found that N2O production decreased significantly under low DO in inactivated sludge and pure water with the sole addition of NH2OH, suggesting that low DO inhibited N2O formation via abiotic NH2OH oxidation. Therefore, in comammox Nitrospira-dominant sludge, low DO decreased N2O production primarily due to the inhibition of low DO to N2O formation via abiotic NH2OH oxidation and the low NH2OH accumulation. These results imply that enriching comammox Nitrospira in the wastewater treatment process can ensure stably low N2O emissions regardless of the variations in DO.

|
Scooped by
?
June 23, 11:09 AM
|
Recombinant proteins play a crucial role in both fundamental research and biotechnology. In the laboratory, recombinant proteins are used in a myriad of ways, including to label cells, localize proteins and isolate complexes. In the clinic, antibody-based therapeutics can dramatically increase cancer survival rates, while virus-like particles (VLPs) are being developed as next-generation vaccines. These innovations have escalated demands for biopharmaceutical recombinant proteins. However, in traditional systems (e.g. mammalian and microbial) the expression of recombinant proteins can be prohibitively expensive. One sustainable, low-cost solution is to use a microalgal-based expression system, such as Chlamydomonas reinhardtii, Phaeodactylum tricornutum, Chlorella sp., Haematococcus pluvialis or Nannochloropsis gaditana. Tools for microalgal protein expression are developing rapidly. Yet our understanding of recombinant protein expression and purification in microalgal systems lags that of traditional systems. Here, we review the impact of commonly used affinity and epitope tags (e.g. Polyhistidine-tag, Strep-tag II, HA-tag and FLAG-tag) on recombinant protein detection, purification and biofunctionality in microalgae. Additionally, we review fluorescent protein tags (such as GFP, mVenus, DsRed and mCherry) and protease cleavage sites, including ‘self-cleaving’ 2A peptides. Finally, we provide guidance on experimental design to enhance the likelihood of successfully expressing recombinant proteins in microalgae.

|
Scooped by
?
June 23, 10:55 AM
|
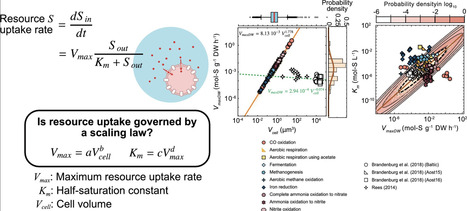
Microbial growth is often described in terms of resource uptake rates, making the understanding and parameterisation of these rate-limiting processes critical for microbial modelling. In phototrophic plankton, theoretical studies suggest that nutrient uptake is limited by mechanistic processes involving membrane transporters, and it has been observed that the cell-specific maximum resource uptake rate (Vmax) follows a power-law relationship with cell size, as well as a trade-off between Vmax and the half-saturation constant (Km). These constraints may also apply to chemotrophic microorganisms; however, many datasets lack direct cell-size measurements. We therefore leveraged the assumption that prokaryotic cell sizes, Vmax, and Km each follow log-normal distributions, drawing parallels with established phytoplankton scaling laws. Our analysis suggests that chemotrophic organisms generally exhibit higher maximum uptake rate per dry weight (VmaxDW) and Km values than phototrophs, and that VmaxDW and Km are not strongly correlated when all chemotroph data are combined. Furthermore, the Bayesian-derived exponents for VmaxDW and Km exceeded those expected from allometric scaling relationships based on the membrane-transport capacity observed for phototrophs, implying that a range of additional factors likely affect observed kinetic parameters.

|
Scooped by
?
June 23, 10:35 AM
|
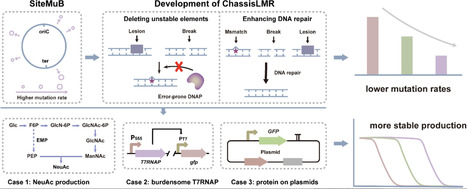
Microbial bioproduction is an important approach to realizing green biomanufacturing. However, poor bioproduction stability caused by genetic heterogeneity is one of the important factors limiting its industrial-scale applications. Here, two methods have been developed to reduce genetic heterogeneity in Bacillus subtilis. SiteMuB (the site-dependent mutation bias) was proposed to enable stable genome integration expression by analyzing the spontaneous mutation rate of the same DNA sequences integrated at different genome sites. Additionally, robustly growing chassis with low mutation rates (ChassisLMR) were developed by deleting unstable elements and enhancing DNA repair. These methods were then employed to improve the production stability of small molecule metabolites and proteins. In N-acetylneuraminic acid production, after 76 generations of cell division, corresponding to the number of cell generations required for > 200-m3 industrial-scale production, strains with SiteMuB and ChassisLMR achieved 15.9-fold and 11.1-fold higher titres than that of the starting strain, respectively. Moreover, by improving the genetic stability of burdensome T7RNAP, combining SiteMuB with ChassisLMR stably maintained the T7 expression system for up to 74 generations, representing a 2.1-fold improvement. Furthermore, ChassisLMR improved the production stability of GFP on the plasmids by 1.38-fold. Overall, SiteMuB and ChassisLMR provide broadly applicable and highly efficient ways to achieve stable bioproduction by reducing genetic heterogeneity.

|
Scooped by
?
June 23, 10:24 AM
|
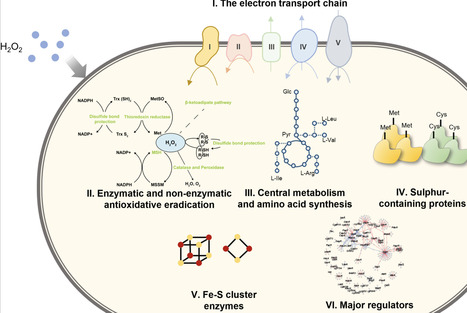
Corynebacterium glutamicum serves as a pivotal industrial chassis for biomanufacturing and an ideal model for studying the phylogenetically related pathogen Mycobacterium tuberculosis. Oxidative stress poses a critical challenge to microorganisms during aerobic industrial processes and immune cell-mediated antibacterial killing by perturbing cellular redox homeostasis, affecting central metabolism, and damaging the integrity of biomacromolecules. However, the intricate mechanisms underlying the dynamic defence of C. glutamicum, despite previous transcriptomic studies on acute and adaptive responses to oxidative stresses, remain largely unclear, hindering strain engineering for industrial applications and the development of effective antimicrobial treatments. In this study, the susceptibility of C. glutamicum to hydrogen peroxide (H2O2) was evaluated, and the inhibitory dynamics of H2O2 were characterized through viable cell counting. RNA sequencing (RNA-seq) was employed to analyse gene expression changes after exposure to 720 mM H2O2. The treatment induced differential expression of 966 and 787 genes at 2 and 6 h, respectively, reflecting perturbations across a broad array of pathways, including (i) enhanced H2O2 and peroxide scavenging, mycothiol biosynthesis, and iron chelation; (ii) repressed central metabolism and enhanced anaplerosis; (iii) elevated sulphur assimilation; (iv) altered amino acid biosynthesis; and (v) altered transcriptional regulation in response to oxidative stress. Further validation by overexpression of ahpD, cysN, and exogenous supplementation with l-methionine and l-cysteine significantly enhanced bacterial tolerance to H2O2. Overall, this study provides the most comprehensive analysis to date of temporal cellular adaptation to H2O2 stress in C. glutamicum, establishing a foundation for future applications in both biomanufacturing and antimicrobial research.

|
Scooped by
?
June 23, 10:11 AM
|
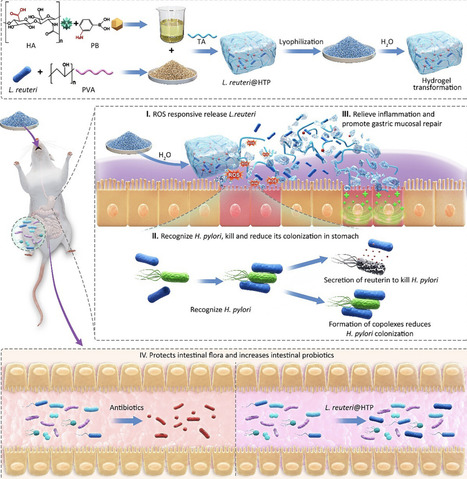
Lactobacillus reuteri (L. reuteri) therapies represent a potentially effective approach to eradicating Helicobacter pylori (H. pylori). However, the difficulty in bacterial viability preservation and harsh gastric environment compromises the survival and on-target delivery of L. reuteri. This study presents a novel bacterium-mediated bacterial elimination strategy using an edible L. reuteri@HTP probiotic powder for targeted bacterial elimination. The probiotic powder is obtained by grinding a lyophilized hydrogel composed of L. reuteri, hyaluronic acid (HA), tannic acid (TA), and polyvinyl alcohol (PVA). Upon contact with water, the powder quickly transforms into a hydrogel, enhancing L. reuteri’s survival in the harsh gastric environment and ensuring selective release at H. pylori-infected inflammatory sites. L. reuteri targets and reduces H. pylori colonization while secreting reuterin to eliminate the bacteria. Additionally, TA's antioxidant properties help alleviate inflammation, and HA supports gastric mucosal repair. L. reuteri@HTP powder preserves the integrity of the gut microbiota, facilitating the restoration of a healthy microbiome. In particular, the probiotic powder remains stable at room temperature for at least six months, providing a promising alternative to traditional antibiotics for H. pylori treatment. This strategy combines targeted eradication, mucosal healing, and microbiome restoration, offering a new approach to treating gastric infections.

|
Scooped by
?
June 23, 9:56 AM
|
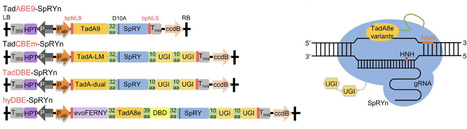
Base editing enables precise nucleotide substitutions within a relatively broad editing window (5–6 nucleotides). However, considerable bystander editing significantly compromise its accuracy. Point mutagenesis, a powerful approach for gradient-tuning protein function, facilitates the generation of diverse plant phenotypes to meet the demands of complex environments and consumer preferences. Here, a series of plant base editors is engineered by fusing three optimized TadA8e variants, TadA9, TadA-LM, and TadA-dual, with a PAM-flexible SpRY nickase (SpRYn, with 5′-NNN PAM recognition). These editors enable A-to-G, C-to-T, and dual-base (simultaneous A-to-G and C-to-T) conversions within a highly condensed active window (1–3 nucleotides). Performance evaluations reveal that the TadDBE (TadA Dual-Base Editor) achieves the most robust outcomes, delivering dual-base editing efficiencies ranging from 2.3% to 61.4%, while maintaining minimal off-target activity. Utilizing TadDBE, targeted point mutagenesis is performed on OsBadh2, a gene encoding betaine aldehyde dehydrogenase that plays a critical role in the biosynthesis of 2-acetyl-1-pyrroline (2-AP), a key aromatic compound. This approach yields rice lines exhibiting gradient-tuned aromatic profiles and optimized levels of 2-AP and γ-aminobutyric acid (GABA). These evolved TadA-derived editors provide a precise, PAM-flexible platform for base editing and represent a versatile strategy for generating genome-edited plants with gradient-tuned agronomic traits.

|
Scooped by
?
June 23, 9:31 AM
|
Risk assessment frameworks for plant agricultural biotechnology products have been in place for decades, focused on the evaluation of living biotechnology products created through genetic engineering. These products contain genetic material from outside the breeder’s gene pool, which is often from different taxa or represents “novel combinations of genetic material”. These products are typically considered to be “genetically modified” (GM) organisms in regulatory jurisdictions. However, in the microbial world, particularly among Bacteria and Archaea, the rapid expansion of genome sequence databases shows that natural microbial innovation primarily occurs through the natural exchange of genetic material from various sources, even from different taxa. This means that many microbes can be considered naturally occurring GM organisms. This raises the question of whether labeling a microbe as GM is always scientifically relevant for risk assessment. In most regulatory frameworks, being classified as GM significantly impacts the registration path, especially for microbes intended for environmental release. A more effective and science-based regulatory approach would assess the actual functions of a microbe rather than relying on the uncertain classification of its genetic material. This would benefit regulators, developers, and society by promoting the use of microbial technologies for agricultural use.
|
 Your new post is loading...
Your new post is loading...