Your new post is loading...
 Your new post is loading...
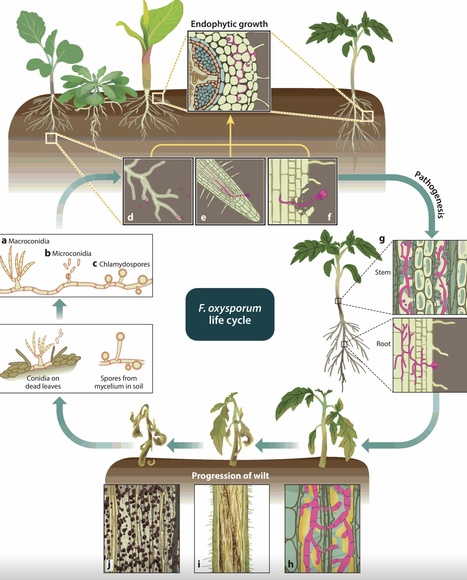
Vascular wilt fungi are a group of hemibiotrophic phytopathogens that infect diverse crop plants. These pathogens have adapted to thrive in the nutrient-deprived niche of the plant xylem. Identification and functional characterization of effectors and their role in the establishment of compatibility across multiple hosts, suppression of plant defense, host reprogramming, and interaction with surrounding microbes have been studied mainly in model vascular wilt pathogens Fusarium oxysporum and Verticillium dahliae. Comparative analysis of genomes from fungal isolates has accelerated our understanding of genome compartmentalization and its role in effector evolution. Also, advances in recent years have shed light on the cross talk of root-infecting fungi across multiple scales from the cellular to the ecosystem level, covering their interaction with the plant microbiome as well as their interkingdom signaling. This review elaborates on our current understanding of the cross talk between vascular wilt fungi and the host plant, which eventually leads to a specialized lifestyle in the xylem. We particularly focus on recent findings in F. oxysporum, including multihost associations, and how they have contributed to understanding the biology of fungal adaptation to the xylem. In addition, we discuss emerging research areas and highlight open questions and future challenges.
Via The Sainsbury Lab
Bioengineering of plant immune receptors has emerged as a key strategy for generating novel disease resistance traits to counteract the expanding threat of plant pathogens to global food security. However, current approaches are limited by rapid evolution of plant pathogens in the field and may lack durability when deployed. Here, we show that the rice nucleotide-binding, leucine-rich repeat (NLR) immune receptor Pik-1 can be engineered to respond to a conserved family of effectors from the multihost blast fungus pathogen Magnaporthe oryzae. We switched the effector binding and response profile of the Pik NLR from its cognate rice blast effector AVR-Pik to the host-determining factor pathogenicity toward weeping lovegrass 2 (Pwl2) by installing a putative host target, OsHIPP43, in place of the native integrated heavy metal–associated domain (generating Pikm-1OsHIPP43). This chimeric receptor also responded to other PWL alleles from diverse blast isolates. The crystal structure of the Pwl2/OsHIPP43 complex revealed a multifaceted, robust interface that cannot be easily disrupted by mutagenesis, and may therefore provide durable, broad resistance to blast isolates carrying PWL effectors in the field. Our findings highlight how the host targets of pathogen effectors can be used to bioengineer recognition specificities that have more robust properties compared to naturally evolved disease resistance genes.
Via The Sainsbury Lab

|
Scooped by
Kamoun Lab @ TSL
August 9, 1:27 PM
|
I spent three days at a leadership course. These are the most helpful takeaways.

|
Scooped by
Kamoun Lab @ TSL
August 9, 1:25 PM
|
Let’s review the year through our team’s publications, blogs and other contributions.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:58 PM
|
These are my opening remarks for the symposium held on November 4th, 2024, in Norwich. This event marks the launch of the strategic partnership between The Sainsbury Laboratory and the Khalifa Center for Genetic Engineering and Biotechnology. Our collaboration aims to advance climate-resilient plant immunity research by uniting our expertise in plant-pathogen interactions specific to desert and dryland plants.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:55 PM
|

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:51 PM
|
A powerful mix of emotions from a family visit, current events, and the incredible success of the Paris Olympic Games brought me to tears. I cried today. When you live your life as a permanent expat, going back home is always an emotional roller coaster. Catching up with my mother, always vigilant for any signs that something might be off. Meeting uncles and aunts who aren’t getting any younger. Spending quality time with my sister and her in-laws. Being there at that special moment when my niece found out she was admitted to med school. It all adds up, leaving you emotionally drained by the end of the week. By the time the week was over, I was emotionally spent. But I didn’t cry. It wasn’t until I boarded the return flight to London and started reading about the Olympics closing ceremony that the tears came, and I couldn’t hold them back.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:48 PM
|
From Neopalpa donaldtrumpi to Syllipsimopodi bideni, let’s stop naming species after racists. An anthem for our times Perhaps no moment better captures the current state of America than Ingrid Andress’ rendition of the US national anthem, “The Star-Spangled Banner,”at a Major League Baseball game on July 15 in Arlington, Texas. Following the performance, Andress admitted to being intoxicated and has since checked herself into rehab. This incident coincides with a contentious presidential election between a convicted felon and a frail and incoherent senior, perfectly reflecting the nation’s turmoil. Predictably, Andress’ music has gone viral after her unforgettable performance reflecting an unhealthy fixation on flawed individuals. Botanists take a stand for a more inclusive future Meanwhile, biologists attending the International Botanical Congress in Madrid have decided to purge more than 200 Latin binomials containing racial slurs related to the root “caffr-” and rename them with the innocuous “affr-” alternative. Latin binomials are scientific names for organisms that use two terms: the genus and the species. We are Homo sapiens, and potato is Solanum tuberosum. You have to thank the Swedish botanist Carl Linnaeus for systematically applying the system back in the 18th century, specifically in 1753 with the publication of “Species Plantarum” and in his subsequent work “Systema Naturae,” but it has been religiously followed ever since. Incidentally, although this naming system is widely known as Linnean nomenclature, it was in fact originally developed by the brothers Gaspard and Johann Bauhin as early as 1622.

|
Scooped by
Kamoun Lab @ TSL
August 30, 2024 7:08 AM
|
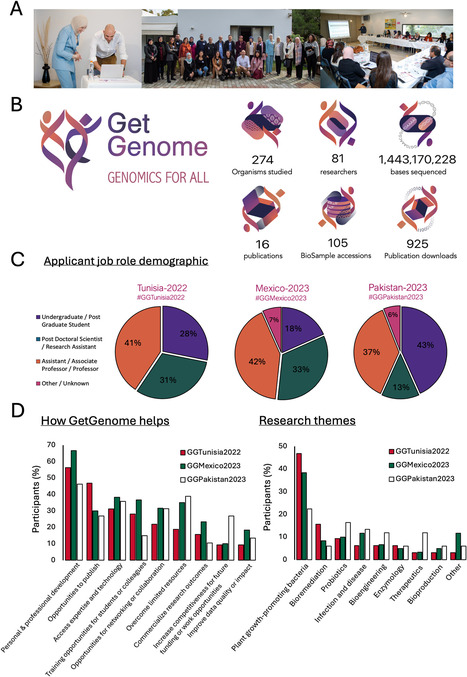
Although genomics has become integral to life science research, access to genomics technology remains inequitable. This article introduces GetGenome, a non-profit organization that aims to overcome this by providing global and affordable access to genomics technology and training.

|
Scooped by
Kamoun Lab @ TSL
August 30, 2024 7:05 AM
|
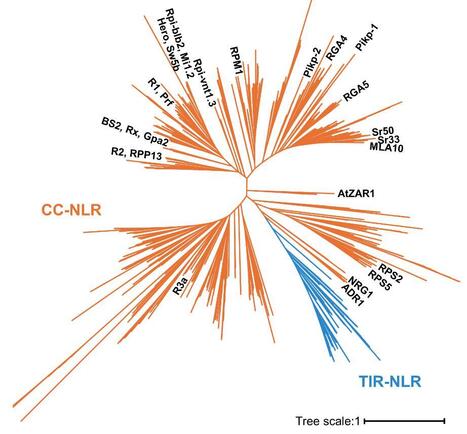
In recent years, the increase in genome sequencing across diverse plant species has provided a significant advantage for phylogenomics studies, allowing the analysis of one of the most diverse gene families in plants: nucleotide-binding leucine-rich repeat receptors (NLRs). However, due to the sequence diversity of the NLR gene family, identifying key molecular features and functionally conserved sequence patterns is challenging through multiple sequence alignment. Here, we present a step-by-step protocol for a computational pipeline designed to identify evolutionarily conserved motifs in plant NLR proteins. In this protocol, we use a large-scale NLR dataset, including 1,862 NLR genes annotated from monocot and dicot species, to predict conserved sequence motifs, such as the MADA and EDVID motifs, within the coiled-coil (CC)-NLR subfamily. Our pipeline can be applied to identify molecular signatures that have remained conserved in the gene family over evolutionary time across plant species.

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:24 AM
|
“As public scientists, we express dismay and sadness at having suffered unjustified destruction, a result of obscurantism and anti-scientific knee-jerk reactions.”

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:21 AM
|
This week, an eclectic group of host-parasite interaction scientists gathered at the immaculate Janelia Research Campus for a conference titled “Mechanisms of Inter-Organismal Extended Phenotypes.” Here are the key takeaways.

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:16 AM
|
For me, writing is all about the vibe; I need to feel that good karma vibe to be creative. This week, I hit a creative roadblock, so I decided to delve into my writing process to overcome it.
|
In plants, NLR (nucleotide-binding domain and leucine-rich repeat) proteins execute innate immunity through the formation of resistosomes that accumulate at the plasma membrane. However, the extent to which NLR resistosomes target other cellular membranes is unknown. Here, we show that the helper NLR NRG1 engages with multiple organellar membranes to trigger innate immunity. Compared to other helper NLRs, NRG1 and closely related RPW8-like NLRs (CCR-NLRs) possess extended N-termini with distinctive sequence signatures, enabling their assembly into longer structures than canonical coiled coil NLR (CC-NLR) resistosomes. Activated NRG1 associates with single- and double-membrane organelles via its N-terminal RPW8-like domain. Our findings reveal that plant NLR resistosomes accumulate at a variety of cellular membrane sites to activate immunity.
Via The Sainsbury Lab

|
Scooped by
Kamoun Lab @ TSL
August 9, 1:29 PM
|
We’re living in dark times. These three short stories reflect that. - The Chumash wale
- Mistaken identity
- Mandela is a terrorist

|
Scooped by
Kamoun Lab @ TSL
August 9, 1:26 PM
|
Costa Rica is home to 5% of the world’s biodiversity. Here are eight fascinating natural history facts we discovered during a visit to this incredible country. Pura vida!

|
Scooped by
Kamoun Lab @ TSL
August 9, 1:22 PM
|
Steve Jobs was obsessed with hiring “the best people.” You should be too. I have always believed that, as a research leader or institution, the most critical decisions we make are about hiring. Bringing the right people into the team can create a ripple effect of benefits, amplifying positive impacts across the lab, the institution, and even the broader research community. These individuals not only contribute directly but also foster an environment of collaboration, innovation, and mutual support that elevates everyone’s work. Conversely, hiring problematic individuals can have a corrosive impact, introducing stress, tension, and conflict that disrupt productivity and morale, often extending far beyond their immediate surroundings. Careful and thoughtful hiring is not just about filling a role — it is about shaping the culture, vision, and long-term success of the team and institution.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:57 PM
|
This week, I committed statistical blasphemy by arguing that, in some cases, one replicate can be enough for a specific type of molecular biology experiment. Before you rush to conclusions, let me explain. I haven’t lost my mind. Click here to listen to the podcast. A single experiment would prove me wrong One of my favorite quotes comes from Albert Einstein: “No amount of experimentation can ever prove me right.” He was likely emphasizing the importance of falsification in science, as he continued, “but one experiment can prove me wrong.” What I like about this quote What I appreciate most about this quote isn’t just the emphasis on falsification — although that is a critical and often overlooked part of experimental science. What really resonates with me is the dismissal of the idea that an experiment can provide a providential answer. Yes, occasionally, a single experiment can be incredibly informative, and yes, there are those Aha moments when one experiment yields a powerful clue to a process. But no experiment happens in a vacuum. It can’t be taken out of the context of how we view the world at that particular moment. That’s the crux of the matter — and it’s where we see the difference between two competing schools of statistics: frequentists vs. Bayesians. To be less and less wrong I’ve touched on Bayesian thinking before in a previous essay, Death by Statistics, so I won’t dwell too much on it here. But essentially, unlike frequentists who treat individual experiments as completely disconnected from any broader context, Bayesians embrace the concept of prior knowledge — or priors for short. The focus isn’t on single experiments in isolation, but on the impact they have on our overall understanding of the world. What matters most is whether an experiment — or any new piece of knowledge — shifts us closer to or further away from the probability that a phenomenon is true. This is especially relevant when new data emerges. Different scientists will start with different levels of belief in a conclusion’s correctness. But the key isn’t where they start; it’s whether, as new evidence comes in, those probabilities inch upward or downward. Ideally, the scientific community moves toward consensus — the famed Bayesian convergence. This is the dream for any field of science: that new knowledge steadily coalesces around a set of robust findings, helping us continually refine our understanding of the natural world.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:53 PM
|
Resurrect Bio is pioneering sustainable agriculture through Disease Resistance Resurrection. By enhancing natural plant defenses of our crops, our London based startup is reducing the need for harmful chemicals and safeguarding global food security. How It started In the ever-evolving field of plant pathology, breakthroughs often come from the most unexpected places. This story begins with Cian Duggan, an ambitious Irish scientist who was wrapping up his PhD under the mentorship of Tolga Bozkurt at Imperial College London. Cian’s research was groundbreaking; he demonstrated that plant immune receptors can accumulate precisely at the sites of pathogen infection — a novel concept that challenged existing paradigms in plant immunity. Cian had also a knack for taking chances and forging his own path. Despite his academic success and the excitement surrounding his findings, Cian harbored an enduring dream: to bridge the gap between academia and industry by founding his own biotech company. He envisioned leveraging his research to develop innovative solutions that could revolutionize agriculture and safeguard global food security.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:50 PM
|
At The Sainsbury Laboratory (TSL), our reputation is built on the stellar research outputs of our scientists. We’re proud to be recognized as a world-leading institute in molecular plant-microbe interactions, but we believe our role extends beyond publishing groundbreaking research. We aspire to be the beating heart of the plant biology community, a vibrant hub where novel knowledge and technologies are rapidly deployed and shared. TSL Summer Conference One of the ways we extend our impact is through the Summer Conference on Plant-Microbe Interactions. This event brings together international keynote speakers, local speakers and selected Early Career Researchers. The event, which extends to two weeks, offers plenty of time for informal and social interactions to discuss the latest approaches and discoveries in Plant-Microbe Interactions.

|
Scooped by
Kamoun Lab @ TSL
December 15, 2024 3:45 PM
|
We owe it to insects. We really do. Without insects, we would have no colorful flowers. Imagine how drab the world would be without flowers. Flowers are the spectacular result of coevolution between plants and insects. When flowers evolved in plants around 130 million years ago, they took advantage of insects to help them pollinate each other, therefore promoting sexual reproduction and enhancing resilience to environmental changes and diseases. The intricate dance between plants and their insect pollinators is one of nature’s most fascinating stories. Flowers evolved bright colors, enticing scents, and sweet nectar to attract insects, while insects, in turn, evolved specialized structures and behaviors to access these rewards and transfer pollen from flower to flower. This mutualistic relationship — plants get insects to transfer pollen between flowers and insects get rewarded with food— has driven an incredible diversification of both plants and insects, filling our world with an astounding array of forms and functions.

|
Scooped by
Kamoun Lab @ TSL
August 30, 2024 7:07 AM
|
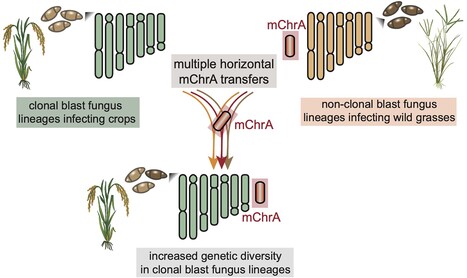
Crop disease pandemics are often driven by asexually reproducing clonal lineages of plant pathogens that reproduce asexually. How these clonal pathogens continuously adapt to their hosts despite harboring limited genetic variation, and in absence of sexual recombination remains elusive. Here, we reveal multiple instances of horizontal chromosome transfer within pandemic clonal lineages of the blast fungus Magnaporthe (Syn. Pyricularia) oryzae. We identified a horizontally transferred 1.2Mb accessory mini-chromosome which is remarkably conserved between M. oryzae isolates from both the rice blast fungus lineage and the lineage infecting Indian goosegrass (Eleusine indica), a wild grass that often grows in the proximity of cultivated cereal crops. Furthermore, we show that this mini-chromosome was horizontally acquired by clonal rice blast isolates through at least nine distinct transfer events over the past three centuries. These findings establish horizontal mini-chromosome transfer as a mechanism facilitating genetic exchange among different host-associated blast fungus lineages. We propose that blast fungus populations infecting wild grasses act as genetic reservoirs that drive genome evolution of pandemic clonal lineages that afflict cereal crops.

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:26 AM
|
Join me on a journey from the breezy shores of Tenerife to the far-reaching effects of climate change and other man-made catastrophes.

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:23 AM
|
Italy launches first field trial for gene-edited crops.

|
Scooped by
Kamoun Lab @ TSL
July 5, 2024 11:20 AM
|
You might not want to hear them, but here they are anyway. - Be generous with your efforts
- Join X/Twitter
- One paper a day keeps the supervisor away
- Back to biology basics
- Learn to structure your thoughts and projects
- Acquire some skills but avoid hyperspecializing
- Make sure you understand Bayesian reasoning
|



 Your new post is loading...
Your new post is loading...