 Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
June 24, 11:22 AM
|
Rhizobia are soil-dwelling proteobacteria that can enter into symbiotic nitrogen-fixing relationships with compatible leguminous plants. Taxonomically, rhizobia are divided into alpha-rhizobia, which belong to the class Alpharoteobacteria, and beta-rhizobia, which belong to the class Betaproteobacteria. To date, all bona fide alpha-rhizobia belong to the order Hyphomicrobiales. However, a recent study suggested that Sphingomonas sediminicola DSM 18106T is also a rhizobium and is capable of nodulating pea plants (Pisum sativum), which would expand the known taxonomic distribution of alpha-rhizobia to include the order Sphingomonadales. Here, we attempted to replicate the results of that previous study. Resequencing and computational analysis of the genome of S. sediminicola DSM 18106T failed to identify genes encoding proteins involved in legume nodulation or nitrogen fixation. In addition, experimental plant assays indicated that S. sediminicola DSM 18106T is unable to nodulate the two cultivars of pea tested in our study, unlike the rhizobium Rhizobium johnstonii 3841T. Taken together, and in contrast to the previous study, these results suggest that S. sediminicola DSM 18106T is not capable of inducing root nodule formation on pea, meaning that the taxonomic distribution of all known alpha-rhizobia remains limited to the class Hyphomicrobiales.

|
Scooped by
Jean-Michel Ané
June 23, 6:03 PM
|
Some species of legumes and nine other flowering plant families form symbioses with bacteria that fix atmospheric nitrogen within specialized plant structures called nodules. How and how often nodulation symbiosis originated has implications for engineering symbiotic nitrogen fixation in non-legume crops. The prevailing hypothesis of a single origin with massive parallel losses has been challenged in a phylogenomic study favoring 16 origins and 10 losses. Nodulation has been assembled once or many times from existing processes (e.g., mycorrhizal symbiosis) and therefore almost nothing about it is truly novel. Because any feature of nodulation can be explained either as divergence from a common origin or as convergence in unrelated taxa, tests are needed that can distinguish whether assembly of homologous components has occurred uniquely or convergently. Much needs to be learned about nodulation symbioses across the proposed independent origins, especially involving the master nodulation transcription factor, Nodule Inception (NIN).

|
Scooped by
Jean-Michel Ané
June 23, 5:49 PM
|
GmbHLHm1 is a basic Helix-Loop-Helix membrane (bHLHm1) DNA binding transcription factor localized to the symbiosome membrane and nucleus in soybean (Glycine max ) nodules. Overexpression of GmbHLHm1 significantly increased nodule number and size, nitrogen fixation activity,and nitrogen delivery to the shoots. This contrasts with reduced nodule numbers per plant, nitrogen fixation activity and poor plant growth when silenced using RNAi. The promoter of GmbHLHm1 was found to be sensitive to exogenous GA supply, decreasing the level of GUS expression in transformed hairy roots in both nodules and roots and reducing native GmbHLHm1 expression in wild-type nodules. In summary, our study suggests that GmbHLHm1 positively regulates soybean nodulation and nitrogen fixation, and that GA can negatively regulate GmbHLHm1 expression in soybean nodules.

|
Scooped by
Jean-Michel Ané
June 23, 11:57 AM
|
• A monoculture of T. obliquus does not grow in nitrogen-free medium.
• Selected bacteria supply nitrogen (N) required for microalga T. obliquus growth due to N-fixation.
• T. obliquus in co-culture with NFB5 had similar metabolic activity in both N-free and complete media.
• NFB5 benefits the growth as T. obliquus as Synechocystis sp. in co-culture.

|
Scooped by
Jean-Michel Ané
June 20, 11:28 AM
|
Our knowledge about how non-coding genetic variation influences phenotypes in animals has deepened considerably in the last decade. However, plant research in this area has lagged behind. To help fill this gap, Robert Schmitz and colleagues studied the impact of cis-regulatory diversity in maize. Using single-cell ATAC-seq and single-nuclei RNA-seq, they profiled 172 inbred lines, creating a dataset that includes over 700,000 nuclei and 33 cell states. Their analyses highlight cis-regulatory elements unique to domesticated maize and the evolutionary impact of transposon activity. Importantly, they identified over 100,000 chromatin accessibility–associated genetic variants (cis-caQTLs), many of which are cell-type-specific and overlap with GWAS variants. STARR-seq was used to test their effects on enhancer activity. Variants in TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP)–binding sites seem to contribute in a major way to variation in chromatin accessibility. Integration of cis-caQTLs, trait variation, and population history shed light on how local adaptation remodeled regulatory networks in specific cellular contexts to modulate flowering time and floral morphology in maize. In the future, it will be interesting to extend these types of analyses to study how genetic variation conditions responses to biotic and abiotic stress in maize and in other key crops.

|
Scooped by
Jean-Michel Ané
June 18, 5:21 PM
|
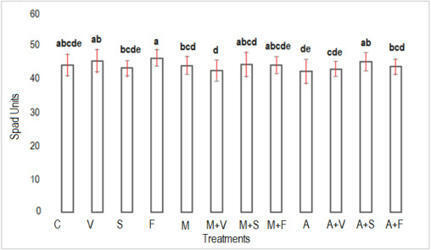
•Biofertilizers promoted the growth and nutrition of pea plants.
•Vermicompost and Azospirillum improved the soil organic matter.
•Mycorrhizal and Azospirillum improved the carbon of the microbial biomass.
•Height and weight improved with mycorrhizal and Azospirillum with vermicompost.
•Azospirrillum and mycorrhiza improved the profitability of the pea crop.

|
Scooped by
Jean-Michel Ané
June 16, 11:59 AM
|
Crops are increasingly exposed to drought and nutrient deficiencies, necessitating enhanced resistance to adverse conditions to meet the growing demands of the global population. While crop productivity has been greatly improved by integrating traits for high yield and stress tolerance through breeding, yield plateaus are now being observed. The rhizosheath, with physical and biological properties distinct from bulk soil, presents a promising target for enhanced tolerance to abiotic stresses such as drought and nutrient deficiencies. This multifunctional region contributes substantially to stress resistance and nutrient cycling, playing a pivotal role in the context of climate change and diminishing supplies of non-renewable fertilisers. We highlight the potential of the rhizosheath as a valuable breeding target to enhance crop productivity under diverse challenging environmental conditions.

|
Scooped by
Jean-Michel Ané
June 15, 10:34 AM
|
We reported that Glycyrrhiza uralensis inoculated with rhizobium tended to increase biomass production and glycyrrhizic acid (GL) production, in this study we have also achieved drastically increase in biomass and GL production in Glycyrrhiza glabra. At thirty days after inoculation (DAI), a significant increase in SPAD values was observed, and the expression of GL synthesis marker genes was also significantly increased. At 150 DAI, a significant increase in biomass was observed. Characteristically, it was also found that thick roots were enlarged by rhizobial inoculation. In addition, the expression of GL synthesis marker genes was also significantly increased. Moreover, GL content per unit root dry weight reached 4%, and GL production per plant increased six times compared to uninoculated plants. Moreover, we tried to reveal the mechanism of induction of GL production by rhizobial inoculation. Since it has been reported that the expression of jasmonic acid (JA) synthesis marker genes is increased by rhizobium in soybean, we investigated the expression of those genes in G. glabra, and found that GgMYC2 and GgJAR1 were up-regulated at Thirty DAI. Furthermore, methyl jasmonate treatment increased the expression of GL synthesis marker genes, suggesting that JA signaling is involved in the increased GL production due to rhizobial inoculation. These results aid in understanding the mechanism of increased GL production through the introduction of rhizobial symbiosis, and show the potential for providing a technology to significantly shorten the cultivation period for the production of Glycyrrhiza that meets the criteria for herbal medicines.

|
Scooped by
Jean-Michel Ané
June 13, 1:16 PM
|
Fungi are one of the most diverse and ecologically important groups of organisms on Earth. They exhibit remarkable diversity in their ecological roles, ranging from decomposers to mutualistic symbionts to parasites. They have a wide array of lifestyles, which reflect their diverse ecological roles and evolutionary adaptations to marine, aquatic, and terrestrial ecosystems. Fungi are osmotrophs that grow as filaments of cells (hyphae) into their food, secrete digestive enzymes across their cells’ chitinous walls, and absorb dissolved nutrients. The classification of fungal lifestyles is primarily based on how they obtain nutrients, with the major modes of nutrition being saprotrophy, parasitism, mutualism and commensalism. Here, we briefly explore these various lifestyles, illustrating their significance in ecosystems and their relationships with other organisms, and then discuss how comparative genomics provides novel insights into their evolutionary trajectories.

|
Scooped by
Jean-Michel Ané
June 13, 11:49 AM
|
Arbuscular mycorrhizal (AM) fungi are ancient plant mutualists that are ubiquitous across terrestrial ecosystems. These fungi are unique among most eukaryotes because they form multinucleate, open-pipe mycelial networks, where nutrients, organelles, and chemical signals move bidirectionally across a continuous cytoplasm. AM fungi play a crucial role in ecosystem functioning by supporting plant growth, mediating ecosystem diversity, and contributing to carbon cycling. It is estimated that plant communities allocate ∼3.93 Gt CO2e to AM fungi every year, much of which is stored as lipids inside the fungal network. Despite their ecological significance, the cellular biology of AM fungi remains underexplored. Here, we synthesise the current knowledge on AM fungal cellular structure and organisation. We examine AM fungal development at different biological levels — the hypha and its content, hyphal networks and AM fungal spores — and explore key cellular dynamics. This includes cell wall composition, cytoplasmic contents, nuclear and lipid organisation and dynamics, network architecture, and connectivity. We highlight how their unique cellular arrangement enables complex cytoplasmic flow and nutrient exchange processes across their open-pipe mycelial networks. We discuss how both established and novel techniques, including microscopy, culturing, and high-throughput image analysis, are helping to resolve previously unknown aspects of AM fungal biology. By comparing these insights with established knowledge in other, well-studied filamentous fungi, we identify critical knowledge gaps and propose questions for future research to further our understanding of fundamental AM fungal cell biology and its contributions to ecosystem health.

|
Scooped by
Jean-Michel Ané
June 13, 11:33 AM
|
Symbiotic nitrogen fixation represents a crucial yet energy-demanding strategy for legumes to survive in nutrient-poor soils. We highlight the multifaceted roles of NSP1 and NSP2 in this symbiosis and propose their function as ‘nutrient-responsive regulators’, integrating environmental signals, physiological status, and nutrient availability, to ensure nodulation occurs only under favorable conditions.

|
Scooped by
Jean-Michel Ané
June 12, 11:19 AM
|
Root nodule symbiosis allows for plant acquisition of reactive nitrogen through fixation of atmospheric molecular dinitrogen by nitrogen-fixing bacteria. Nodulation is a complex trait, with diverse modes of bacterial infection and nodule morphologies across species, reflecting evolutionary adaptation. Understanding ancient forms of this trait may carry advantages for its current utilization, since basal states likely reflect the least complexity. In this review we focus on the evolution of nodule development, particularly on events that have led to increased complexity of this symbiosis in later adaptations. We hypothesize that the ancestral form of nodulation comprises of an evolutionary coupling of nutrient-dependent lateral root development with apoplastic intercellular bacterial growth, alongside the acquisition or evolution of an ancestral chitinaceous signaling molecule by the microbial symbiont. Uncovering the evolutionary adaptations underpinning the extant diversity of this trait allows for a better understanding of the simplest ancestral state.

|
Scooped by
Jean-Michel Ané
June 12, 9:45 AM
|
Arbuscular mycorrhiza (AM) with soilborne Glomeromycota fungi was pivotal in the conquest of land by plants almost half a billion years ago. In flowering plants, it is hypothesized that AM is initiated by the perception of AM fungi-derived chito- and lipochito-oligosaccharides (COs/LCOs) in the host via Lysin Motif Receptor-Like Kinases (LysM-RLKs). However, it remains uncertain whether plant perception of these molecules is a prerequisite for AM establishment and for its origin. Here, we made use of the reduced LysM-RLK complement present in the liverwort Marchantia paleacea to assess the conservation of the role played by this class of receptors during AM and in CO/LCO perception. Our reverse genetic approach demonstrates the critical function of a single LysM-RLK, MpaLYKa, in AM formation, thereby supporting an ancestral function for this receptor in symbiosis. Binding studies, cytosolic calcium variation recordings and genome-wide transcriptomics indicate that another LysM-RLK of M. paleacea, MpaLYR, is also required for triggering a response to COs and tested LCOs, despite being dispensable for AM formation. Collectively, our results demonstrate that the perception of symbionts by LysM-RLK is an ancestral feature in land plants, and suggest the existence of yet-uncharacterized AM fungi signals.
|

|
Scooped by
Jean-Michel Ané
June 23, 6:09 PM
|
Phosphate transporters play a key role in improving crop yield. In this study, TaPT31-7A is a high-affinity phosphate transporter strongly induced in arbuscular-mycorrhizal (AM) wheat roots. It restores Pi uptake in yeast mutant MB192 and localizes to the plasma membrane. TaPT31-7A overexpression lines accumulated more shoot and root phosphorus than the wild type under both low- and high-Pi conditions. When inoculated with AM in Pi-deficient soil, these overexpression lines displayed enhanced Pi uptake, higher mycorrhization, and improved growth, ultimately increasing the spikelet number per spike, spike length, 1000-grain weight, grain length, and grain width. Transcriptome and coexpression analyses of TaPT31-7A OE lines and control plants showed altered expression of phosphate-starvation and AM-development genes, while docking and yeast two-hybrid assays confirmed its interaction with PP2C phosphatase TaPP2C12-6A. These results establish TaPT31-7A as a central regulator of Pi uptake, AM symbiosis, and productivity in wheat and highlight its potential for breeding phosphorus-efficient cultivars.

|
Scooped by
Jean-Michel Ané
June 23, 5:56 PM
|
Unlike many fungi, arbuscular mycorrhizal (AM) fungi have proven recalcitrant to genetic manipulation due to their obligate biotrophic lifestyle and multinucleate, coenocytic cellular structure. In this review, we examine past attempts at AM fungal transformation, we identify key biological and technical barriers and explore recent advances to overcome them. We focus on techniques never before applied in AM fungi, including CRISPR/Cas9, microinjection, and protoplast-based transformation, and we explore how they provide viable strategies for achieving this elusive goal. We conclude by outlining guidelines for future research, distinguishing between established approaches that are readily applicable to AM fungi and others that first require addressing key outstanding questions in AM fungal cell biology and genetics to ensure success.

|
Scooped by
Jean-Michel Ané
June 23, 5:47 PM
|
Purine nucleotides are key metabolites of plants because they are constituents of nucleic acids and participate in energy and nitrogen metabolism. They are produced by de novo synthesis or salvage pathways, the latter being energetically favorable. By using CRISPR/Cas9 gene down-regulation along with omics technologies in common bean, López et al. (2025) unveiled functional specialization of two isoforms of adenine phosphoribosyltransferase (APRT) impacting the fate of adenine. Notably, APRT1 played a prevalent role in adenine salvage in roots, whereas APRT5 is involved in cytokinin homeostasis and stress responses. Furthermore, experiments with transgenic hairy roots showed that nodules with down-regulated APRT5 have a considerably larger size and weight than wild-type nodules, indicating a previously unknown function of some APRTs in regulating nodulation.

|
Scooped by
Jean-Michel Ané
June 23, 11:54 AM
|
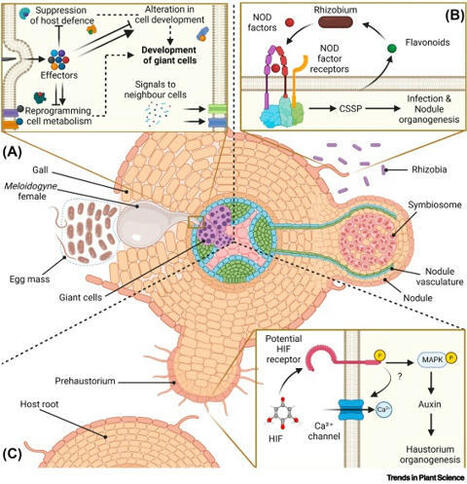
Plants have evolved diverse adaptations in signal perception, hormone regulation, and organ development that enable the formation of specialised structures such as nematode-induced galls, rhizobia-induced nodules, and host-induced parasitic plant haustoria that facilitate both parasitic and mutualistic symbiosis. Despite their differences, these organs share common gene regulatory mechanisms with lateral root development. By comparing their mechanisms of hormonal regulation, we illuminate the shared genetic underpinnings and how plants repurpose vegetative development pathways in response to biotic stimuli. This adaptive retooling positions plants along the symbiotic spectrum from exploited hosts to mutualistic partners and strategic predators. Comparative analysis of the hormonal mechanisms that drive symbiotic organogenesis highlights the plasticity of developmental processes and the interplay between internal signalling and external environmental cues.

|
Scooped by
Jean-Michel Ané
June 18, 5:23 PM
|
Arbuscular mycorrhizal (AM) symbiosis is an important biological breakthrough which assisted plant land colonization over 400 million years ago. This widespread mutualistic interaction between fungi and plants enhances nutrient exchange, ecological sustainability, plant stress resistance, and host plant development. AM symbiosis improves plant nutrition by deriving nutrients through both mycorrhizal pathways and the Plant's own pathways. AMF influence nutrient availability by altering soil properties, microbial populations, and nutrient cycling. Understanding the life cycle of AMF, spore germination, sporulation, colonization, and symbiosis formation are critical for large-scale agricultural applications. Root organ culture (ROC) techniques offer intriguing possibilities to mass producing AMF under in vitro. This review surveys the literature on these topics, focusing on methods for enhancing sporulation in in vitro. Enhancing in vitro sporulation can be achieved by supplementing growth media with phenolic compounds, fatty acids, and phytohormones, and optimizing the media and related factors. These compounds regulate fungal growth and development, leading to increased sporulation and improved AMF inoculant efficacy. Further research is needed to provide quality inoculum and develop crop-specific formulations and delivery methods to harness the potential of AMF in diverse agroecosystems.

|
Scooped by
Jean-Michel Ané
June 17, 2:16 PM
|
Unraveling the mechanisms underlying the maintenance of species diversity is a central pursuit in ecology. It has been hypothesized that ectomycorrhizal (EcM) in contrast to arbuscular mycorrhizal fungi can reduce tree species diversity in local communities, which remains to be tested at the global scale. To address this gap, we analyzed global forest inventory data and revealed that the relationship between tree species richness and EcM tree proportion varied along environmental gradients. Specifically, the relationship is more negative at low latitudes and in moist conditions but is unimodal at high latitudes and in arid conditions. The negative association of EcM tree proportion on species diversity at low latitudes and in humid conditions is likely due to more negative plant-soil microbial interactions in these regions. These findings extend our knowledge on the mechanisms shaping global patterns in plant species diversity from a belowground view.

|
Scooped by
Jean-Michel Ané
June 16, 11:54 AM
|
Globally, invasive plants, animals, and microbes are a dominant threat to biodiversity and ecosystems, inflicting over $1 trillion USD in damage annually. Hundreds of microbial invasive taxa are documented.
Currently, we are inoculating microbes into many environments in enormous numbers to fertilize agricultural soils, remediate contamination, control eutrophication, precipitate minerals, and restore ecosystem functions.
Given the astronomical numbers and myriad environments that are being inoculated with microbes, these manipulations risk creating microbial invasions in much the same way that catastrophic plant and animal invasions have been precipitated by intentional introductions by humans.
A mechanistic understanding and predictive framework for the potential for microbial inoculants to cause invasions is needed to balance their benefits with their risks of causing harmful effects.

|
Scooped by
Jean-Michel Ané
June 15, 10:32 AM
|
Plants and microbiomes have co-evolved for millennia. Through this co-evolution, microbiomes have become essential for plant nutrient acquisition, which involves plant signaling, microbial sensing, and acquiring and sharing nutrients. In this review, we synthesize recent advancements in the complex associations of molecular, physiological, and eco-evolutionary mechanisms that underpin microbe-facilitated plant nutrient uptake. Focusing on emerging insights in plant-microbial communication and metabolic pathways, we evaluate potential opportunities to harness plant microbiomes to sustainably supply nutrients in agricultural and natural ecosystems. However, further progress is constrained by key knowledge gaps. We propose an amended conceptual framework for advancement that includes a holistic understanding of eco-evolutionary relationships with explicit consideration of signaling and sensing mechanisms. Finally, we argue that advancing fundamental science by utilizing emerging analytical approaches in an integrated way is critical to develop effective microbiome-informed tools that can enhance plant nutrient acquisition and promote long-term food security and environmental sustainability.

|
Scooped by
Jean-Michel Ané
June 13, 12:59 PM
|
Breakthroughs in DNA sequencing have upended our understanding of fungal diversity. Only ∼155,000 of the 2–3 million fungal species on the planet have been formally described and named, and ‘dark taxa’ — species known only from sequences — represent the vast majority of species within the fungal kingdom. The International Code of Nomenclature requires physical type specimens to officially recognize new fungal species, making it difficult to name dark taxa. This is a significant problem for conservation because, without names, species cannot be recognized for environmental and legal protection. Symbiotic ectomycorrhizal (EcM) fungi play a particularly important role in forest carbon drawdown, but at present we have little understanding of how many EcM fungal species exist, or where to prioritize research activities to survey and describe EcM fungal lineages. In this review, we use global soil metabarcoding databases (GlobalFungi and the Global Soil Mycobiome consortium) to evaluate current estimates of the total number of EcM fungal species on Earth, outline the current state of undescribed EcM dark taxa, and identify priority regions for future dark taxa exploration. The metabarcoding databases include up to 219,730 EcM fungal operational taxonomic units (OTUs) detected from almost 39,500 samples. Using Chao richness estimates corrected for extrapolating species numbers from metabarcoding datasets, we predict that the global diversity of EcM fungi could be ∼25,500–55,500 species. Dark taxa — those that do not match species-level identities — account for 79–83% of OTUs. Oceania contains the highest percentage of dark taxa (87%), and Europe the lowest (78%). Priority ‘darkspots’ for future research occur predominantly in tropical regions, but also in selected temperate forests at both southern and northern latitudes. We propose concrete steps to reduce the prevalence of EcM darkspots, including performing targeted field surveys, barcoding fungaria voucher specimens, and developing new ways to describe and conserve fungal taxa from DNA alone.

|
Scooped by
Jean-Michel Ané
June 13, 11:45 AM
|
Plants accommodate diverse microbial communities, termed the microbiome, which can change dynamically during plant adaptation to varying environmental conditions. However, the direction of these changes and the underlying mechanisms driving them, particularly in crops adapting to the field conditions, are not well understood. Here, we investigate the root endosphere microbiome of rice (Oryza sativa ssp. japonica) across four consecutive cultivation seasons in a high-yield, non-fertilized, and pesticide-free paddy field, compared to a neighboring fertilized and pesticide-treated field. Using 16S rRNA amplicon and metagenome sequencing, we analyzed three Japonica cultivars—Nipponbare, Hinohikari, and Kinmaze. Our findings reveal that the root endosphere microbiomes diverge based on fertilization regime and plant developmental stages, while the effects of cultivar variation are less significant. Machine learning model and metagenomic analysis of nitrogenase (nif) genes suggest enhanced nitrogen fixation activity in the non-fertilized field-grown roots, highlighting a potential role of diazotrophic, iron-reducing bacteria Telmatospirillum. These results provide valuable insights into the assembly of the rice root microbiome in nutrient-poor soil, which can aid in managing microbial homeostasis for sustainable agriculture.

|
Scooped by
Jean-Michel Ané
June 12, 11:24 AM
|
NifA, the activator of nitrogenase, is sensitive to ammonium concentration, particularly within its N-terminal domain. In this work, genetically engineered mutants with N-terminal deletions of the nifA1 and nifA2 genes were constructed using overlap extension PCR to reduce the inhibitory effect of ammonium on nitrogenase expression in Rhodobacter capsulatus SB1003. Under 3 mM NH4+, the hydrogen production rate of ZX03 (nifA1-, nifA2-) reached 0.65 mmol L−1 h−1, with a 31.2 % increase in hydrogen production compared to the wild-type. When 8 mM NH4+ was used as the sole nitrogen source, H2 production in all strains decreased substantially compared to 5 mM NH4+. However, ZX03 demonstrated a 5.2-fold enhancement in hydrogen production relative to the wild-type under 8 mM NH4+, underscoring its improved ammonium tolerance. During hydrogen production, the gene expression levels of nifA and nifH in all mutant strains were significantly up-regulated under ammonium conditions compared to the wild-type. These findings reveal distinct roles of nifA1 and nifA2 in ammonium tolerance and hydrogen production.

|
Scooped by
Jean-Michel Ané
June 12, 9:49 AM
|
Many plants and animals, including humans, host diverse communities of microbes that provide many benefits. A key challenge in understanding microbiomes is that the species composition often differs among individuals, which can thwart generalization. Here, we argue that the key to identifying general principles for microbiome science lies in microbial metabolism. In the human microbiome and in other systems, every microbial species must find ways to harvest nutrients to thrive. The available nutrients in a microbiome interact with microbial metabolism to define which species have the potential to persist in a host. The resulting nutrient competition shapes other mechanisms, including bacterial warfare and cross-feeding, to define microbiome composition and properties. We discuss impacts on ecological stability, colonization resistance, nutrient provision for the host, and evolution. A focus on the metabolic ecology of microbiomes offers a powerful way to understand and engineer microbiomes in health, agriculture, and the environment.
|





 Your new post is loading...
Your new post is loading...











Interesting review on old players