Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
January 19, 5:41 PM
|
Arbuscular mycorrhizal fungi (AMF) are widespread plant symbionts that enhance nutrient acquisition and influence ecosystem productivity. Previous chromosome-level assemblies of a model species revealed a two-compartment genome architecture (active A and repressed B chromatin compartments), yet its conservation across evolutionarily distant AMF lineages remains unresolved. Here, we present a chromosome-scale and 3D genome assembly of Gigaspora margarita isolate BEG34—the largest and most repeat-rich AMF genome to date—alongside that of its obligate endobacterium, Candidatus Glomerobacter gigasporarum (CaGg), using PacBio HiFi and Hi-C sequencing. The G. margarita genome comprises 43 chromosomes (792 Mb) organized into stable A/B compartments and Topologically Associating Domains structures, irrespective of the presence of endobacteria. We uncover 21 divergent rDNA operons distributed across six chromosomes and show that these physically interact, suggesting conserved nucleolar organization. We also reveal that the CaGg genome is tripartite and mobilome-rich, encoding prophages, an orphan CRISPR array, and complete pathways for many novel and essential cofactors, including heme, which may enhance host bioenergetics. We also find that the endobacterium’s presence regulates transposable elements in G. margarita. These findings reveal conserved principles of chromatin architecture in AMF symbionts and highlight the tight molecular interplay between fungal hosts and their endosymbionts, offering new insights into genome evolution and symbiotic adaptation.

|
Scooped by
Jean-Michel Ané
January 19, 5:34 PM
|
Nitrogen-fixing microbes are a primary contributor of this important nutrient to the global nitrogen cycle. Biological nitrogen fixation (BNF) through the enzyme nitrogenase requires extensive energy that in whole cells is generally studied during the oxidation of carbohydrates such as sugars. The nitrogen-fixing bacterium Azotobacter vinelandii is a model diazotroph for the study of aerobic BNF. Much is known about metabolism in A. vinelandii when cultured on a simple medium where energy is provided primarily in the form of sucrose or glucose. Outside of the laboratory, this soil bacterium grows on metabolites primarily derived from plant root exudates or from the degradation of dead plant matter. In this work, we expand on previous studies looking at genes that are essential to BNF in A. vinelandii when grown on sucrose medium using transposon sequencing (Tn-seq). We applied Tn-seq to determine the genes essential to growth when the medium was shifted to acetate, succinate or glycerol as the primary carbon and energy source to fuel both growth and BNF. A global overview of the genes of central metabolism and those directing substrates toward central metabolism, along with a selection of unexpected genes that were essential for specific growth substrates, is provided.

|
Scooped by
Jean-Michel Ané
January 14, 5:12 PM
|
Agricultural sustainability is becoming increasingly threatened by climate change and anthropogenic activities. Arbuscular mycorrhizal fungi (AMF) provide an important solution through their symbiotic associations with plant roots, enhancing nutrient acquisition, improving soil structure, mitigating stress effects, and contributing to carbon sequestration through glomalin production. AMF improves the bioavailability of essential nutrients by extending hyphal networks and activating nutrient transporter genes. They also aid in heavy metal detoxification by sequestering metals within hyphae, reducing plant uptake, and safeguarding tissues, while supporting carbon sequestration by stabilizing soil aggregates. Across diverse taxa, AMF exhibit species-specific adaptations to different soil types and agroecosystems, contributing to phosphorus solubilization, nitrogen use efficiency, and metal sequestration. Molecular insights revealed that AMF symbiosis activates genes related to nutrient exchange and stress adaptation, including the Ca2⁺ signaling pathways. Advanced inoculation techniques, such as high-efficiency spore propagation, seed pelleting, and bioreactor-based production, provide scalable and contamination-free applications that reduce reliance on synthetic fertilizers and promote sustainable cropping systems. Future research should focus on integrating molecular tools, precision agriculture, and real-time monitoring to optimize AMF application strategies, enhance nutrient cycling, and strengthen soil health, thereby advancing sustainable food systems and addressing global agricultural challenges.

|
Scooped by
Jean-Michel Ané
January 10, 7:17 PM
|
Background
Since the first description of an Azospirillum-like bacterium in 1925 by Martinus Beijerinck in The Netherlands, this genus has become a cornerstone of plant–microbe interactions and sustainable agriculture worldwide. Over the past century, Azospirillum has been extensively studied for its ability to promote plant growth, enhance stress tolerance, and contribute to nutrient acquisition, particularly in cereals and legumes. These functions are mediated by multiple mechanisms, including nitrogen fixation, phytohormone production, and the modulation of the root architecture by effector molecules.
Scope
This review presents a comprehensive synthesis of the historical and current research on Azospirillum and its impact in agriculture and beyond. It explores its taxonomic expansion, physiological versatility, genetic manipulation, and interactions with plant hosts and other microorganisms. It also examines its agronomical impact on extensive and intensive cropping systems, both individually and mixed in microbial consortia. Advances in formulation technologies and regulatory frameworks for commercial inoculants are discussed, as well as cutting-edge tools such as artificial intelligence and multi-omics integration that are reshaping how we understand and deploy this bacterium. Beyond agriculture, Azospirillum has proven valuable in environmental contexts such as revegetation of degraded lands, bioremediation of contaminated soils, and ecological restoration in arid zones. Its capacity to colonize diverse hosts, survive extreme conditions, and contribute to ecosystem processes underscores its potential far beyond agriculture.
Conclusion
One hundred years after its first scientific mention, Azospirillum remains not only relevant but also vital. As the world moves toward more sustainable agricultural and ecological systems, this review reaffirms the legacy and promise of this genus.

|
Scooped by
Jean-Michel Ané
January 8, 2:06 PM
|
Aspergillus fumigatus is a notorious pathogenic fungus responsible for various harmful, sometimes lethal, diseases known as aspergilloses. Understanding the gene regulatory networks that specify the expression programs underlying this fungus’ diverse phenotypes can shed mechanistic insight into its growth, development, and determinants of pathogenicity. We used eighteen publicly available RNA-seq datasets of Aspergillus fumigatus to construct a comprehensive gene regulatory network resource. Our resource, named GRAsp (Gene Regulation of Aspergillus fumigatus), was able to recapitulate known regulatory pathways such as response to hypoxia, iron and zinc homeostasis, and secondary metabolite synthesis. Further, GRAsp was experimentally validated in two cases: one in which GRAsp accurately identified an uncharacterized transcription factor negatively regulating the production of the virulence factor gliotoxin and another where GRAsp revealed the bZip protein, AtfA, as required for fungal responses to microbial signals known as lipo-chitooligosaccharides. Our work showcases the strength of using network-based approaches to generate new hypotheses about regulatory relationships in Aspergillus fumigatus. We also unveil an online, user-friendly version of GRAsp available to the Aspergillus research community.

|
Scooped by
Jean-Michel Ané
January 5, 9:24 PM
|
Rhizobial technology has become a transformative tool for environmentally friendly and sustainable agriculture. Rhizobia are key nitrogen-fixing bacteria that enhance soil fertility and reduce reliance on synthetic nitrogen fertilisers. In addition to nitrogen fixation, they act as effective plant growth promoters by producing phytohormones, mobilising nutrients, and improving root development. Advances in bioinoculant engineering now support efficient symbiotic associations in both leguminous and non-leguminous crops, offering a green strategy to boost agricultural productivity. Rhizobia also help plants withstand abiotic and biotic stresses, and many strains display strong biocontrol abilities by producing antimicrobial compounds and suppressing phytopathogens. However, their field performance can be inconsistent due to poor survival during storage, competition with native microbes, environmental conditions, and limited farmer awareness. To overcome these challenges, strategies such as co-inoculation with compatible microbes, encapsulated formulations, genetic enhancement, improved agronomic practices, pathogen management, and farmer awareness are being developed to increase inoculant stability and effectiveness. Overall, rhizobial technology serves as a cornerstone of smart, sustainable farming, supporting food security, environmental protection, and the restoration of soil health for future green agriculture.

|
Scooped by
Jean-Michel Ané
January 4, 8:50 PM
|
Modern agriculture faces an urgent need to improve nutrient use efficiency while reducing environmental impacts. Here, we show that ancestral traits controlling rhizosphere microbiome functions can be reintroduced into elite maize through targeted teosinte introgressions. Using near-isogenic lines, we mapped microbiome-associated phenotypes (MAPs) derived from teosinte that suppress nitrification and denitrification—key microbial processes contributing to nitrogen loss. These introgressions altered root exudate chemistry, resulting in distinct microbial assemblies and enhanced nitrogen retention. We identified candidate loci and exudate metabolites responsible for suppressive activity and demonstrated their functional effects in vitro. These findings reveal a genetic and biochemical basis for rewilding microbiome-mediated ecosystem services in crops, offering a scalable path toward sustainable nutrient management in global agriculture.

|
Scooped by
Jean-Michel Ané
January 2, 4:41 PM
|
Cytokinin (CK) is a key regulator of root system architecture including primary root (PR) and lateral root (LR) development. During root nodule symbiosis in legumes, CK promotes nodule organogenesis in the root cortex while simultaneously suppressing infection thread (IT) formation in the epidermis, yet the molecular mechanisms enabling this spatial specificity remain incompletely understood. TCP INTERACTOR CONTAINING EAR MOTIF PROTEIN (TIE) proteins negatively regulate CK signaling in Arabidopsis roots, but whether legume orthologs modulate symbiotic CK responses remains unknown.
We characterized Lotus tie1 loss-of-function mutants through root and nodulation assays, transcriptomics, confocal microscopy of CK signaling reporter TCSn, and ethylene quantification.
LjTIE1 expression was induced by CK and Nod-factor, tie1 mutants exhibited reduced PR length, LR density, IT formation, and nodule number alongside elevated ethylene emission - phenotypes fully rescued by ethylene biosynthesis inhibition. Despite enhanced CK signaling (confirmed via TCSn reporter and transcriptomics), tie1 roots formed no spontaneous nodules but displayed hypernodulation under exogenous CK treatment.
LjTIE1 provides temporal control of symbiosis by dampening CK signaling after nodule initiation, revealing a regulatory layer that prevents constitutive nodulation despite elevated CK perception.

|
Scooped by
Jean-Michel Ané
December 31, 2025 4:11 PM
|
Soil, the Earth's upper crust layer, is crucial for ecological processes, comprising mineral, organic, and biological components that determine fertility and multifuncionality. Human-induced degradation necessitates advancements in pedology and soil conservation. The rhizosphere, surrounding plant roots, houses a diverse microbial community, notably bacteria, which enhance plant growth and disease resistance. Root exudates fuel biological activity and nutrient cycling, supporting microbial growth, improving soil structure, and reducing plant stress. Plant-microorganism interactions in ecological and agricultural systems play a vital role for maintaining primary production and ecosystem sustainability. Moreover, arbuscular mycorrhizae and nitrogen-fixing bacteria are essential, influencing plant development, sustainability, and ecosystem health. Specific bacterial phyla populate the rhizosphere and endosphere, with Plant Growth-Promoting Rhizobacteria (PGPR), such as Pseudomonas spp. and Bacillus spp., playing a prominent role. PGPR employ direct and indirect mechanisms, including phytohormone production, mineral solubilization, systemic resistance induction, antibiosis, competition for resources, and ACC deaminase activity, The amalgamation of these traits underscores the conceptual foundation for comprehending the ecological and agricultural implications of employing microbes. This inquiry is particularly relevant to sustainable agriculture, where the use of microbes, including PGPR, plays a crucial role in biofertilization and mitigating environmental stressors. Thus, investigating the ecological and agricultural implications through multi-omics approaches such as genomics, transcriptomics, proteomics, and metabolomics offers valuable insights. The integration of these multi-omics data provides a comprehensive framework for understanding the complex interactions between plants, bacteria, and fungi. This holistic perspective not only deepens our understanding of soil ecology but also lays the groundwork for informed and sustainable agricultural practices, fostering resilience against environmental stresses.

|
Scooped by
Jean-Michel Ané
December 31, 2025 3:50 PM
|
Microbiota-mediated nutrient turnover in the rhizosphere determines nutrient bioavailability, thereby enhancing nutrient uptake, utilization, and ultimately crop productivity. Consequently, elucidating the functional core microbiota in rhizosphere nutrient turnover is of critical importance. In this study, we leveraged soybean germplasm core collections to investigate the tripartite relationship among host genotype, core microbiota and nutrient availability, with a focus on delineating the pivotal role of core microbiota in nutrient turnover. Our results suggest that phylogenetic variation significantly shape root-associated microbial communities and rhizosphere nutrient availability, explaining 11.75 % and 2.07 % of total variances, respectively. Core microbiota analysis identified 29 phylogenetic conserved core amplicon sequence variants (ASVs), the majority of which exhibited significant correlated with nutrient availability. Notably, three key core ASVs—ASV13, ASV14 and ASV12, positively correlated with alkali-hydrolyzed nitrogen, available phosphorus, and soil organic matter, respectively. These taxa were subsequently incorporated into a Bradyrhizobium-based synthetic bacterial community (SynCom) to validate their functional roles. Further experiments confirmed that core microbiota-driven nutrient turnover directly facilitates host plant, as evidenced by SynCom inoculation assays. Collectively, this study establishes that phylogenetically conserved core microbiota critically regulate nutrient turnover and acquisition efficiency in the rhizosphere. These insights advance our understanding the ecological function of core microbiota in the rhizosphere and provide a framework for harnessing the beneficial traits in sustainable agriculture.

|
Scooped by
Jean-Michel Ané
December 28, 2025 5:02 PM
|
Transmission electron microscopy was the key for revealing structural similarities between intracellular plant-microbe interactions.

|
Scooped by
Jean-Michel Ané
December 25, 2025 9:43 PM
|
Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs that rely on host-derived symbiotic carbohydrates. However, it remains unclear whether symbiotic AMF can access exogenous non-symbiotic carbon sources, complicating our understanding of their relationship with host plants. Here, we investigated the direct uptake of exogenous 13C1-labeled myristate by three symbiotic AMF species (Rhizophagus irregularis, R. intraradices, and R. diaphanous) and assessed their growth responses using AMF-carrot hairy root co-culture systems. Furthermore, we explored the environmental distribution of myristate, and evaluated the impact of exogenous myristate on the carbon-phosphorus exchange between R. irregularis and alfalfa or rice in a greenhouse experiment. Symbiotic AMF can absorb exogenous myristate, as evidenced by 13C enrichment and transcriptional activation of fatty acid transport and metabolism genes in AMF extraradical hyphae. Myristate is commonly present in various soil and plant environments, and its application increased both intraradical and extraradical fungal biomass, possibly linked to suppressed mycorrhizal-activated defense responses in host roots. Unexpectedly, exogenous myristate reduced the mycorrhizal phosphorus benefits for both alfalfa and rice and decreased their symbiotic carbon allocation to root-colonizing AMF, although these effects varied with soil phosphorus conditions. These findings provide new insights into understanding and manipulating the nutritional interactions between AMF and host plants.

|
Scooped by
Jean-Michel Ané
December 25, 2025 9:27 PM
|
Crop rotation enhances agroecosystem sustainability by reducing nutrient loss, improving soil fertility, and decreasing crop evapotranspiration. Arbuscular mycorrhizal fungi (AMF) inoculation further supports enhanced nutrient and water uptake by plants, potentially improving water use efficiency and soil health while reducing fertigation needs. However, crop species may respond differently to AMF inoculation under varying fertigation regimes. In this study, the response of four horticultural species to AMF inoculation was investigated under optimal (100 %) and deficit (25 % reduction) water and fertilizer (fertigation) availability. Leek, courgette, white bean, and celery were planted consecutively over the course of two years, with crop production and quality as well as leaf nutrients and soil parameters being measured at the end of each crop cycle. Mycorrhizal inoculation did not improve any agronomic parameter among the crops studied under either fertigation condition.
|

|
Scooped by
Jean-Michel Ané
January 19, 5:39 PM
|
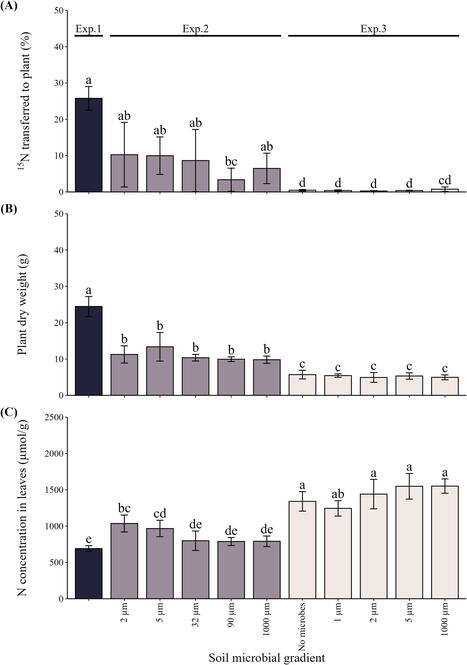
Bacground: Arbuscular mycorrhizal (AM) fungi enhance plant nutrient acquisition from soil; however, their ability to exploit organic nutrient forms in the absence of associated microbes capable of mineralization remains unclear.
Methods: To test if the AM fungi carry their beneficial bacterial partners into nutrient-rich zones, we conducted three controlled experiments manipulating the microbial inputs, diversity and composition in plant–AM fungus–soil systems, ranging from open pots to semi-sterile mesocosms. We manipulated soil microbial diversity by imposing a microbial diversity gradient (complex communities fractionated by size, resulting in fractions passing through 1 µm to 1000 µm sieves) and cultivated Andropogon gerardii in previously sterilized substrate together with a bacterial-free Rhizophagus irregularis. In each experiment, 15N‐labeled chitin or mineral nitrogen (N) compartments were installed in the root‐free zone of each mesocosm.
Results: With decreasing microbial inputs into the root-free zone, the N uptake from chitin to plants, facilitated by the AM fungal hyphae, decreased. Upon complete absence of microbes in the root-free zone, AM hyphal foraging preferences assessed by quantitative PCR indicated that exploration of the mineral N compartments was more effective than that of the chitin compartments. The AM fungal hyphae were ineffective in priming mineralization of organic N even if provided with complex soil microbiomes at a distance from the compartment.
Conclusions: In summary, chitin-enriched compartments become attractive for the AM fungi only when previously mineralized by competent microbes. Such microbes, however, were not effectively transported to spatially restricted organic resources in soil via AM hyphal highways in our experiments.

|
Scooped by
Jean-Michel Ané
January 17, 9:47 PM
|
Bacterial pathogens and most nitrogen-fixing rhizobia employ type III effectors (T3Es) as potent tools to manipulate plant signaling pathways, thereby facilitating infection and colonization. However, how rhizobial T3Es regulate legume symbiosis remains elusive. Here, we show that NopM, a T3E from Sinorhizobium fredii NGR234, contributes to infection and nodulation in Lotus japonicus Gifu. The loss of nopM in an NGR234ΔnopT mutant reduced infection and nodulation in L. japonicus, and expression of NopM under the control of L. japonicus NIN promoter enhanced these processes. NopM associated with the NF receptors NFR1 and NFR5 and physically interacted with their cytosolic domains in vitro, and selectively mediated ubiquitination of NFR5. Expression of NopM in hairy roots of NFR5-HA transgenic plants correlated with increased NFR5 protein abundance relative to the inactive NopM variant. Taken together, our work suggests that NopM-dependent effects on symbiosis are associated with increased NFR5 abundance, expanding our understanding of rhizobial T3E functionality and the co-evolution of legume-rhizobium symbiosis.

|
Scooped by
Jean-Michel Ané
January 12, 12:46 PM
|
The specific partnership between legumes and rhizobia relies on a chemical dialogue. Plant flavonoids activate the bacterial transcription factor NodD, which triggers production of Nod factors that are recognized by the plant. Structural studies of the Pisum sativum (pea) symbiont Rhizobium leguminosarum NodD revealed two pockets that are essential for its activation by flavonoids. Comparative studies with NodD1 of Sinorhizobium medicae, the symbiont of Medicago truncatula, revealed that this specificity is determined by the shape of the pocket and by specific amino acids. A chimeric NodD containing the flavonoid recognition residues from S. medicae NodD1 in the R. leguminosarum NodD backbone was sufficient to complement nitrogen fixation in M. truncatula by an S. medicae nodD1 mutant, confirming the critical role of flavonoid recognition in host range.

|
Scooped by
Jean-Michel Ané
January 9, 7:30 PM
|
During host-microbe symbioses, the fitness of mutualistic microbes is determined by the interactions that concurrently occur, throughout their life cycle, with their host and other members of the surrounding microbial community. Disentangling how these multiple interactions shape the fitness of microbial symbionts is challenging, but is essential to understand the diversity and functioning of mutualisms. Here we examined the different fitness components of rhizobial symbionts of the legume plant Mimosa pudica across the multiple stages of their symbiotic life cycle. By comparing rhizobial symbiotic fitness in single and pairwise inoculations, we found that inter-bacterial interactions causing significant fitness effects are common, transitive and can have major consequences, sometimes leading to the extinction of a strain. These interactions predominantly occur at the root infection (nodulation) step, but smaller post-infection interaction effects, involving yet uncharacterized mechanisms, were also detected. Furthermore, considering pairwise interactions was sufficient to predict fitness ranks in more complex rhizobial communities consisting of 6 or 8 strains, indicating that higher-order interaction effects do not play a significant role in these communities. Overall, our results provide a quantitative framework to describe the main drivers of rhizobial symbiotic fitness in a simple community context.

|
Scooped by
Jean-Michel Ané
January 8, 11:38 AM
|
The Definitive Handbook of Azospirillum" stands out for its comprehensive and specialized focus on Azospirillum, its detailed analysis of the evolutionary knowledge, and its practical experimental proposals. It offers a detailed examination of Azospirillum, while other texts tend to provide broader, more general, or technical perspectives on related topics.
This book not only explores the biological and ecological significance of Azospirillum but also covers the various roles it plays in different environments, especially in agriculture where it has potential applications as a biofertilizer. By providing both historical context and cutting-edge research, this book bridges the gap between past discoveries and future possibilities, making it a valuable tool for researchers and professionals.

|
Scooped by
Jean-Michel Ané
January 5, 3:48 PM
|
The root nodule symbiosis of plants with nitrogen-fixing bacteria is phylogenetically restricted to a single clade of flowering plants, which calls for as yet unidentified trait acquisitions and genetic changes in the last common ancestor. Here we discovered—within the promoter of the transcription factor gene Nodule Inception (NIN)—a cis-regulatory element (PACE), exclusively present in members of this clade. PACE was essential for restoring infection threads in nin mutants of the legume Lotus japonicus. PACE sequence variants from root nodule symbiosis-competent species appeared functionally equivalent. Evolutionary loss or mutation of PACE is associated with loss of this symbiosis. During the early stages of nodule development, PACE dictates gene expression in a spatially restricted domain containing cortical cells carrying infection threads. Consistent with its expression domain, PACE-driven NIN expression restored the formation of cortical infection threads, also when engineered into the NIN promoter of tomato. Our data pinpoint PACE as a key evolutionary invention that connected NIN to a pre-existing symbiosis signal transduction cascade that governs the intracellular accommodation of arbuscular mycorrhiza fungi and is conserved throughout land plants. This connection enabled bacterial uptake into plant cells via intracellular support structures such as infection threads, a unique and unifying feature of this symbiosis.

|
Scooped by
Jean-Michel Ané
January 2, 4:43 PM
|
Alfalfa (Medicago sativa L.), known as “Queen of forages”, is valued to its high-nutritional quality and is a key member of Leguminosae family. Its productivity is largely attributed to mutualistic symbioses with arbuscular mycorrhizal fungi (AMF) and rhizobia, which facilitate nutrient exchange and plant growth. However, the coexistence and mutualistic interactions between rhizobia and AMF across alfalfa genotypes with differing yields in native soil remain poorly understood. In this study, we investigated the community composition of rhizobia and AMF colonizing alfalfa roots across different-yield varieties. Our results showed variations in dominant microbial taxa and the structural complexity of root-associated microbial networks among genotypes. Moreover, rhizobia exhibited no significant associations with AMF on genus level, however, negative correlations were observed among genera within the AMF community, and a comparable trend was identified among rhizobial taxa. In summary, our findings offer new insights into how native soil microbiota influence the dual symbiotic relationships of alfalfa, with implications for leveraging native microbial communities to enhance sustainable forage production.

|
Scooped by
Jean-Michel Ané
December 31, 2025 4:13 PM
|
Arbuscular mycorrhizal (AM) associations of plants and Glomeromycotina soil fungi play a crucial role in all terrestrial ecosystems. In this mutually beneficial interaction, obligate biotrophic fungi acquire photosynthetically fixed carbon from the plant, while the mutualistic fungi enhance plant access to soil nutrients. AM fungi colonize the inner tissues of host roots, where they form specialized symbiotic structures (arbuscules) within fully differentiated cortex cells that are reprogrammed to host the microbe. Given the intimate nature of the interaction, extensive partner communication at the interface of plant and fungal cells is crucial for the development and functioning of AM symbiosis. The peri-arbuscular space, a specialized apoplast compartment surrounding the arbuscules, supports not only nutrient exchange between the symbiotic partners but is also the site of extensive partner crosstalk mediated by cell wall components, receptors, signaling peptides, and extracellular vesicles. Such signaling processes in the apoplast modulate plant immune responses to enable colonization by beneficial fungi, making this compartment a key player for the establishment and maintenance of AM symbiosis. In this review, we discuss recent discoveries related to the role of partner communication in the apoplast, with a focus on peptide and cell wall signaling, as well as extracellular vesicles.

|
Scooped by
Jean-Michel Ané
December 31, 2025 4:10 PM
|
Ectomycorrhizal fungi form symbiotic relationships with a wide range of terrestrial plants, acquiring carbohydrates for themselves and promoting nutrient uptake in their host plants. However, some ectomycorrhizal fungi cannot effectively obtain the thiamine necessary for growth from their host or synthesize it themselves. Ectomycorrhizal fungi can recruit hypha-associated microorganisms, which play a vital role in promoting nutrient absorption and ectomycorrhizal root formation, ultimately colonizing within fruiting bodies to form a unique bacterial microbiota. In this study, non-targeted metabolomics and whole-genome sequencing were employed to investigate the colonization characteristics of the hyphae-associated bacterium Bacillus altitudinis B4 on the mycelial surface of ectomycorrhizal fungus Suillus clintonianus, as well as the synergistic promotion of thiamine synthesis and absorption by B. altitudinis B4 and the fungal mycelium, respectively. The results suggested that S. clintonianus first secreted ureidosuccinic acid and pregnenolone, recruiting the hyphae-associated bacterium B. altitudinis B4 to the mycelial surface. Subsequently, the ureidosuccinic acid secreted by S. clintonianus further stimulated B. altitudinis B4 to enhance thiamine production by increasing its biomass and upregulating the expression of related functional genes. Finally, S. clintonianus absorbed the thiamine secreted by the B. altitudinis B4, promoting fungal growth and increasing the colonization rate in association with Pinus massoniana. This study elucidates the thiamine acquisition mechanisms of ectomycorrhizal fungi, highlighting the critical role of bacterial partners in fungal nutrition and host-fungal interactions.

|
Scooped by
Jean-Michel Ané
December 31, 2025 3:43 PM
|
In the current context of climate change, there is a need to develop more sustainable agrifood strategies. As an alternative to the intensive use of chemically synthesized fertilizers and pesticides that pollute water and impact biodiversity, there is a growing interest in using beneficial microbes as biostimulants and/or bioprotection agents. However, their implementation in agriculture remains a challenge due to highly variable outcomes and benefits. Furthermore, there are major knowledge gaps about the molecular mechanisms that regulate different plant–microbe interactions. In the present review, we summarize current knowledge on the molecular mechanisms that control different beneficial plant root–microbe interactions; namely, arbuscular mycorrhiza, the rhizobium–legume symbiosis, ectomycorrhiza, and fungal and bacterial endophytic associations. This includes the signaling pathways required for recognition of microbes as beneficial, the metabolic pathways that provide nutritional benefits to the plant, and the regulatory pathways that modulate the extent of symbiosis establishment depending on soil nutrient availability and plant needs. Our aim is to highlight the main common mechanisms, as well as knowledge gaps, in order to promote the use of microbes, either individually or in consortia, within the framework of a sustainable agriculture that is less dependent on chemicals and more protective of biodiversity and water resources.

|
Scooped by
Jean-Michel Ané
December 28, 2025 4:57 PM
|
PerCon SFA project data dentification of spatially resolved biomarkers of drought in Sorghum bicolor rhizosphere molecular-microbe interactions using a novel root cartography "RhizoGrid" system for sampling plants under drought and control conditions across 10 equally sized root zone environments (4 quadrants each). Each quadrant was sampled and processed for 16S amplicon, metabolomics, and X-ray computed tomography (XCT). Data download includes experimental metadata and results files for 16S rRNA sequence analysis of microbial community assembly (processed data files), liquid chromatography mass spectrometry (LC-MS) metabolomics analysis of microbial community root exudates (processed data files), X-ray computed tomography (XCT) spatial gradient analysis (raw and processed data files) of microbial community composition, and related computational modeling outputs.

|
Scooped by
Jean-Michel Ané
December 25, 2025 9:34 PM
|
Background and aims
Mycorrhizal (AMF) benefits to ancient tetraploid and hexaploid wheats, particularly under saline conditions, are not sufficiently known.
Methods
A two-year field experiment and a pot experiment were carried out, where the field experiment encompassed non-saline and saline (120 mM NaCl) irrigation, presence and absence of AMF (Funneliformis mosseae) inoculation, and 10 wheat genotypes. The pot experiment encompassed four salinities (0, 40, 80, and 120 mM) and two levels of AMF inoculation (with and without of AMF inoculation) and 11 genotypes.
Results
Salinity suppressed the chlorophyll, carotenoids, K, and P, grains/m2, grain yield, harvest index, and dry mass. Though, it boosted the activities of antioxidative enzymes, Na, electrolyte leakage, Na/K, protein, wet gluten, and gluten index. Inoculation to AMF led to enhancement in the maximum quantum efficiency of photosystem II, chlorophyll, K, P, N, and total phenolic compounds concentrations, the activities of antioxidative enzymes, grains/m2, grain yield, dry mass, protein, wet gluten, and gluten index, while decreasing the Na concentration, Na/K, and electrolyte leakage, particularly in the salt-stricken plants; favorable responses to the AMF were more appreciable in the salt-stricken modern wheats, than the ancient emmer and spelt wheats.
Conclusion
Salinity and AMF exerted contrasting effects on physiological, growth, dry mass, and grain yield attributes of different genotypes, with a tendency of salt-induced suppressions and AMF-induced enhancements to be less notable in the ancient wheats, than the modern bread and durum wheats. Though, salinity and AMF inoculation shared a same trend in improving the grain and flour quality attributes.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...