Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 6:03 PM
|
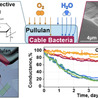
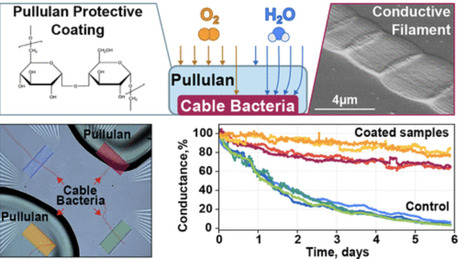
The greening of electronics remains a grand societal challenge, with no radical improvement within sight. Sustainable solutions for electronics, such as biobased and transient materials, are hence receiving growing attention. Presently, there are no biobased alternatives to conventional conductors such as metals and organic polymers, as their conductivity is too low. The discovery of cable bacteria, which are filamentous microorganisms capable of conducting electricity over centimeter-scale distances, has the potential to change this. In cable bacteria, conductivity occurs through thin wires embedded in the cell envelope, displaying conductivities comparable to those of the best highly doped organic polymers. However, exposure to ambient air leads to a gradual loss of their conductivity. To enhance stability, a bioderived protective coating could be useful, thus retaining a fully biobased system. To this end, we investigated pullulan, a polysaccharide polymer primarily used in food packaging that is known for its excellent oxygen-barrier properties. Cable bacterium filaments protected with a film derived from a 10 wt % pullulan solution exhibited a 10-fold increase in conduction stability under ambient conditions compared to uncoated controls. Reducing ambient moisture also preserved the long-term conductivity of the cable bacteria, even in the absence of a protective coating, indicating that humidity plays a critical role in conductance deterioration. Our findings provide an important step toward further technological implementation of the highly conductive wires of cable bacteria and offer practical guidelines for developing biobased coatings for O2-sensitive materials in electronics, thus contributing to the advancement of next-generation green technologies.

|
Scooped by
mhryu@live.com
Today, 5:58 PM
|
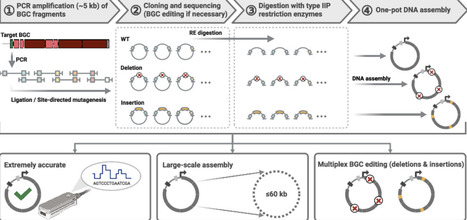
Natural products (NPs) produced by actinobacteria, particularly Streptomyces species, represent a rich source of bioactive compounds and have yielded many clinically important compounds. Actinobacterial genomes are characterized by high GC content and typically harbor 20–40 biosynthetic gene clusters (BGCs) per genome, which encode diverse NPs such as polyketides, peptides, and glycosides. CRISPR/Cas-based genome editing has emerged as a promising tool to activate silent BGCs and engineer NP biosynthesis. However, the efficiency of multiplex editing drastically decreases as the number of targeted sites increases. Here, we report a novel one-pot DNA assembly method, termed direct pathway synthesis and editing (DiPaSE), for the efficient synthesis and multiplex editing of long, high-GC BGCs. DiPaSE accurately assembles multiple high-GC DNA fragments up to 60 kb and enables simultaneous deletions and insertions within a target BGC without compromising the assembly efficiency. Using this approach, we identified functions of previously uncharacterized genes in the aureothin BGC and significantly enhanced the titer of the corresponding NP. The workflow employs conventional polymerase chain reaction, type IIP restriction enzymes, commercially available DNA assembly reagents, and E. coli, providing a simple, cost-effective, and broadly applicable platform for genome mining, BGC refactoring, and rational design of artificial biosynthetic pathways.

|
Scooped by
mhryu@live.com
Today, 5:46 PM
|
Dye pollution in water poses serious health and ecological risks, requiring wastewater treatment before discharge and prompting increased research attention due to the widespread use of dyes in various industries. This study investigates the biosorption of methylene blue (MB) using a novel composite of chitosan and Bacillus subtilis exopolysaccharides (EPS). Fourier-transform infrared spectroscopy (FTIR) analysis confirmed the presence of essential functional groups for dye adsorption. The biosorption process was pH-dependent, with optimal removal efficiencies at pH 6 for the chitosan/EPS composite and pH 7 for chitosan alone, showing increased adsorption capacity with rising pH from 3.0 to 7.0. Contact time experiments demonstrated efficient MB removal in approximately 30 min, achieving decolorization rates of 71.6% for the composite and 60.62% for chitosan. The composite also demonstrated a higher maximum biosorption capacity (14.26 mg g−1) than chitosan (13.70 mg g−1) according to the Langmuir isotherm model, which best described the monolayer adsorption process. Kinetic analyses revealed that the pseudo-second order model best described the adsorption process, with calculated capacities closely matching experimental values (41.67 mg g−1 for chitosan and 45.87 mg g−1 for the composite). Post-adsorption FTIR results revealed involvement of –OH, –NH, –COO−, and –PO₄³− groups via electrostatic, hydrogen-bonding, and π–π interactions, confirming the interaction between MB and functional groups on the biosorbents. These findings highlight the synergistic effect of combining chitosan with EPS, resulting in a promising, efficient, and rapidly equilibrating biosorbent for mitigating dye pollution in wastewater. This work presents the first reported use of Bacillus subtilis exopolysaccharide combined with chitosan as a fully biodegradable composite biosorbent, exhibiting superior adsorption capacity (14.26 mg g−1), markedly faster kinetics (K₂ = 0.176 g mg−¹ min−¹), and equilibrium attainment within 30 min compared to pristine chitosan and most reported biopolymer-based adsorbents. Optimizing biosorption conditions can enhance dye removal efficiencies and contribute to sustainable environmental management practices.

|
Scooped by
mhryu@live.com
Today, 5:36 PM
|
Hepatitis C virus (HCV) and many other RNA viruses contain a type IV internal ribosome entry site (IRES) in their 5′ untranslated region (UTR). These IRES RNAs interact directly with the ribosome, enabling cap-independent translation initiation. Using bioinformatic homology searches, we identify a putative type IV IRES within the annotated 3′ UTR of megrivirus E (MeV-E). In addition to its unusual genomic location, the MeV-E 3′ IRES has a reduced size compared with HCV and many type IV IRESs. We determine the 3D structure of the MeV-E 3′ IRES in complex with the ribosome using cryoelectron microscopy (cryo-EM) and show that the MeV-E 3′ IRES initiates translation, but at lower levels than the larger IRES in the MeV-E 5′ UTR. This small type IV IRES enables translation of a second open reading frame in the MeV-E genome, which likely encodes a transmembrane protein conserved in other megriviruses.

|
Scooped by
mhryu@live.com
Today, 1:18 PM
|
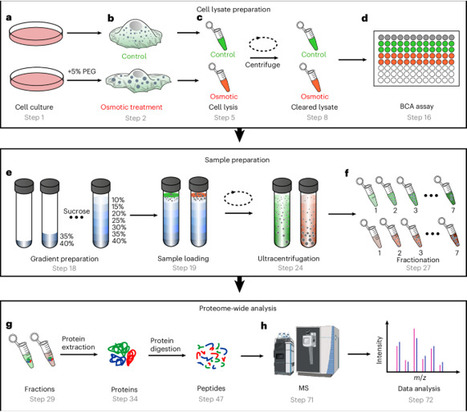
Biomolecular condensates formed through liquid–liquid phase separation regulate cellular processes, and their dysregulation causes disease. Current methods for identifying endogenous phase-separating proteins have low throughput and cannot capture dynamic responses to stimuli. Here we present a protocol combining osmotic compression or transforming growth factor-β (TGF-β) treatment to induce condensation with sucrose density gradient centrifugation and quantitative mass spectrometry to enable systematic, high-throughput identification of endogenous condensates and phase-separating proteins. The method exploits the density changes that occur when phase-separating proteins undergo oligomerization during condensate formation. In H1975 cells, we identified over 1,500 phase-separating proteins under osmotic compression or TGF-β treatment; 538 of these candidates were not present in PhaSepDB, a database that compiles in vivo, in vitro and omics-derived proteins. The approach detects constitutive condensates and proteins that dynamically phase-separate in response to osmotic stress or TGF-β signaling. This protocol provides proteome-wide analysis of fractions of proteins having different densities and enables temporal resolution of phase-separation events. The procedure takes ~9 d and requires expertise in cell culture, biochemistry and mass spectrometry. This method enables systematic study of biomolecular condensates and disease-associated phase-separation mechanisms. This protocol identifies endogenous biomolecular condensates by combining sucrose density gradient centrifugation with quantitative mass spectrometry, enabling proteome-wide, stimulus-responsive profiling of phase-separating proteins.

|
Scooped by
mhryu@live.com
Today, 12:54 PM
|
Thermogenetics enables noninvasive spatiotemporal control over protein activity in living cells and tissues, yet its applications have largely been restricted to transcriptional regulation and membrane recruitment. Here, we present a generalizable strategy for engineering thermosensitive allosteric proteins through the insertion of optimized Avena sativa LOV2 domain variants. Applying this approach to a diverse set of structurally and functionally unrelated proteins in E. coli, we generated potent, thermoswitchable chimeric variants that can be tightly controlled within narrow temperature ranges (37–41 °C). Extending this strategy to mammalian systems, we engineered CRISPR–Cas genome editors directly modulated by subtle temperature changes within the physiological range. Lastly, we showcase the incorporation of a chemoreceptor domain as an alternative thermosensing module, suggesting thermosensitivity to be a widespread feature in receptor domains. This work expands the toolkit of thermogenetics, providing a blueprint for temperature-dependent control of virtually any protein of interest. Thermogenetics enables spatiotemporal control of protein activity using temperature. Now, engineering of a compact, insertable thermoresponsive protein module diversifies the classes of proteins amenable to allosteric thermoregulation.

|
Scooped by
mhryu@live.com
Today, 11:59 AM
|
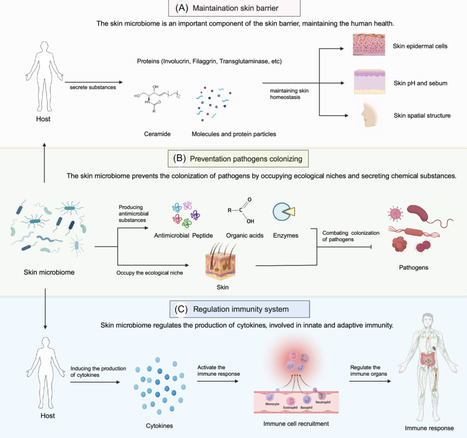
The skin microbiome, consisting of a vast array of microorganisms, is essential for human skin health, aiding in barrier protection, immune regulation, wound repair, and defense against pathogens. Disruptions in this microbial balance are closely linked to the onset and worsening of various skin disorders. This review evaluates the potential of skin microbiome engineering as a therapeutic strategy for treating skin diseases. We discuss nontargeted approaches like probiotics and fecal microbiota transplantation that aim to reshape the microbial community, as well as targeted methods such as phage therapy, phage lysins, and engineered bacteria, which specifically modulate microbial populations or influence the skin environment. These approaches open new avenues for personalized dermatological treatments. Despite significant progress, challenges remain in the clinical translation of microbiome-based therapies. Safety, standardization, regulatory approval, and long-term ecological stability must be addressed to ensure efficacy and reproducibility in clinical settings, underscoring the critical need for further research in their dermatological applications.

|
Scooped by
mhryu@live.com
Today, 11:51 AM
|
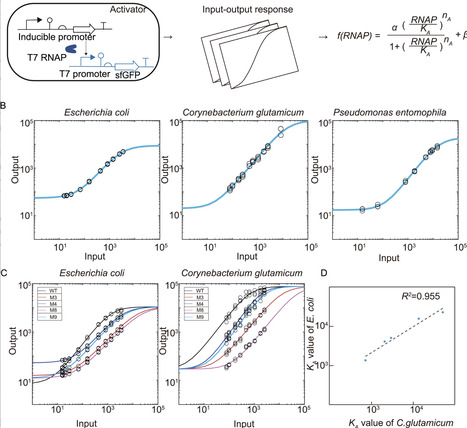
Although the principles of synthetic biology were initially established in model bacteria, microbial producers, extremophiles and gut microbes have now emerged as valuable prokaryotic chassis for biological engineering. Extending the host range in which designed circuits can function reliably and predictably presents a major challenge for the concept of synthetic biology to materialize. In this work, we systematically characterized the cross-species universality of two transcriptional regulatory modules—the T7 RNA polymerase activator module and the repressors module—in three non-model microbes. We found striking linear relationships in circuit activities among different organisms for both modules. Parametrized model fitting revealed host non-specific parameters defining the universality of both modules. Lastly, a genetic NOT gate and a band-pass filter circuit were constructed from these modules and tested in non-model organisms. Combined models employing host non-specific parameters were successful in quantitatively predicting circuit behaviors, underscoring the potential of universal biological parts and predictive modeling in synthetic bioengineering.

|
Scooped by
mhryu@live.com
Today, 11:39 AM
|
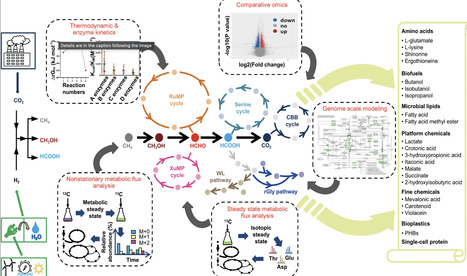
Developing methylotrophic cell factories that can efficiently catalyze organic one-carbon (C1) feedstocks derived from electrocatalytic reduction of carbon dioxide into bio-based chemicals and biofuels is of strategic significance for building a carbon-neutral, sustainable economic and industrial system. With the rapid advancement of RNA sequencing technology and mass spectrometer analysis, researchers have used these quantitative microbiology methods extensively, especially isotope-based metabolic flux analysis, to study the metabolic processes initiating from C1 feedstocks in natural C1-utilizing bacteria and synthetic C1 bacteria. This paper reviews the use of advanced quantitative analysis in recent years to understand the metabolic network and basic principles in the metabolism of natural C1-utilizing bacteria grown on methane, methanol, or formate. The acquired knowledge serves as a guide to rewire the central methylotrophic metabolism of natural C1-utilizing bacteria to improve the carbon conversion efficiency, and to engineer non-C1-utilizing bacteria into synthetic strains that can use C1 feedstocks as the sole carbon and energy source. These progresses ultimately enhance the design and construction of highly efficient C1-based cell factories to synthesize diverse high value-added products. The integration of quantitative biology and synthetic biology will advance the iterative cycle of understand–design–build–testing–learning to enhance C1-based biomanufacturing in the future.

|
Scooped by
mhryu@live.com
Today, 11:30 AM
|
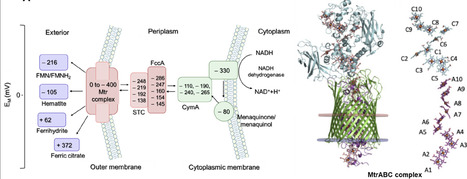
Electroactive microorganisms (EAMs) could utilize extracellular electron transfer (EET) pathways to exchange electrons and energy with their external surroundings. Conductive cytochrome proteins and nanowires play crucial roles in controlling electron transfer rate from cytosol to extracellular electrode. Many previous studies elucidated how the c-type cytochrome proteins and conductive nanowires are synthesized, assembled, and engineered to manipulate the EET rate, and quantified the kinetic processes of electron generation and EET. Here, we firstly overview the electron transfer pathways of EAMs and quantify the kinetic parameters that dictating intracellular electron production and EET. Secondly, we systematically review the structure, conductivity mechanisms, and engineering strategies to manipulate conductive cytochromes and nanowire in EAMs. Lastly, we outlook potential directions for future research in cytochromes and conductive nanowires for enhanced electron transfer. This article reviews the quantitative kinetics of intracellular electron production and EET, and the contribution of engineered c-type cytochromes and conductive nanowire in enhancing the EET rate, which lay the foundation for enhancing electron transfer capacity of EAMs.

|
Scooped by
mhryu@live.com
Today, 11:06 AM
|
genome editing: BASIS, bacterial artificial chromosome (BAC) stepwise insertion synthesis; BGM, B. subtilis genome; CasHRA, Cas9-facilitated Homologous Recombination Assembly; CONEXER, conjugation coupled with programmed excision for enhanced recombination; GC, guanine-cytosine; GENESIS, GENomE Stepwise Interchange Synthesis; HAnDy, Haploidization-based DNA Assembly and Delivery in yeast; IWe, inchworm elongation; MRA, meiotic recombination-mediated assembly; NA, not applicable; REXER, Replicon EXcision Enhanced Recombination; SwAP-In, switching auxotrophies progressively for integration; TAR, transformation-associated recombination; YLC, yeast life cycle.

|
Scooped by
mhryu@live.com
Today, 10:00 AM
|
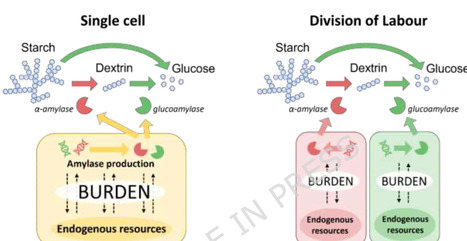
Yarrowia lipolytica is a promising host for a range of biotechnological applications. However, the costs and sustainability of common feedstocks limit the commercial viability of current bioproduction. Consolidated bioprocessing using plant biomass offers a desirable alternative to reduce costs and improve sustainability, though such approaches are often constrained in single-organism systems due to metabolic burden. Here, we have investigated the use of division of labor (DOL) in Y. lipolytica consortia for the degradation of starch, a common component of plant biomass. We engineered a panel of strains expressing α-amylase and/or glucoamylase, with varying promoter and signal peptide combinations. We found that stronger expression led to higher metabolic burden, and that expressing both amylases imposed a greater burden than expressing either enzyme alone. When combining both amylase strains, we found that some consortia could achieve faster growth rates than the equivalent monoculture. The fastest growing consortium identified from the panel was scaled-up into flasks, where the consortium achieved the highest final biomass, although the fastest growth rate was achieved by the α-amylase strain alone. These findings suggest that DOL is a promising strategy for degrading complex macromolecules in plant biomass, offering a potential route to more sustainable bioprocessing.

|
Scooped by
mhryu@live.com
Today, 1:27 AM
|
Glycogen phosphorylase (GP) plays a central role in glycogen metabolism. While the structure and regulation of mammalian GPs have been extensively studied, the corresponding mechanisms in gut bacterial GPs remain poorly understood. Here, we investigate GPs from E. coli (EcGP), Segatella copri (ScGP), and Dorea longicatena (DlGP), which represent three phylogenetic clades of GPs, using enzymatic assays, cryo–electron microscopy (cryo-EM), and X-ray crystallography. We find that ScGP forms a unique pentamer that undergoes adenosine monophosphate (AMP)-dependent assembly into a dimer-of-pentamer, which inhibits activity by restricting substrate access to the catalytic site. EcGP exists in equilibrium among monomers, dimers, and tetramers, with AMP promoting tetramer dissociation and enhancing catalytic efficiency. In contrast, DlGP remains predominantly monomeric and is unresponsive to AMP. These findings uncover structural and regulatory diversity among gut bacterial GPs. Notably, the oligomeric states of GPs modulate substrate accessibility and enzyme activation, suggesting a distinct mode of allosteric regulation beyond the canonical T-to-R transition model. Because bacterial GPs contribute to the generation of glucose, their regulation may influence the composition of gut-derived metabolites that affect host glucose homeostasis and insulin sensitivity. Our study provides mechanistic insight into the structural and functional diversity of gut bacterial GPs and lays a foundation for future exploration of microbiome-mediated metabolic interactions.
|

|
Scooped by
mhryu@live.com
Today, 6:01 PM
|
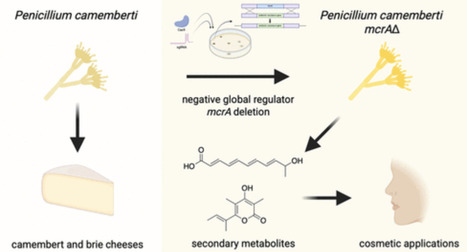
Fungal secondary metabolites have historically provided important applications in a variety of industries. Penicillium camemberti, a fungus with a role in cheese production, was domesticated to food use partly due to its metabolically depleted characteristic, minimizing the risk of toxic compound formation. However, antiSMASH analysis of the genome reveals that strains of the species do contain various cryptic biosynthetic gene clusters and, thus, have the potential capability of producing multiple secondary metabolites despite its limited compound production under normal laboratory conditions. Here, we genetically engineered Penicillium camemberti strain IMV00769, which is genetically similar to cheese-making isolates, by deleting negative global regulator, mcrA. This deletion resulted in the production of secondary metabolites not previously produced by this strain, including fumigermin, a compound patented for cosmetic applications for the reduction of skin wrinkles, enhancement of skin elasticity, and skin whitening. Our findings highlight the power of global regulator manipulation to activate cryptic biosynthetic pathways and expand the range of natural products accessible from domesticated fungal strains.

|
Scooped by
mhryu@live.com
Today, 5:50 PM
|
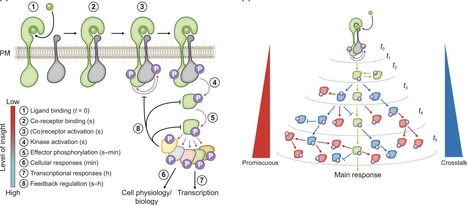
Plants are masters of perception, reacting to a myriad of biochemical and physical cues in a constantly changing environment. Plants rely on local cell-based signal processing to perceive and react sufficiently fast to a multitude of stimuli. The ability to respond quickly is crucial for sustaining growth, defense, and metabolism and thereby the ability to survive challenges associated with pathogens, resource limitations, environmental fluctuations, and mechanical perturbations. Protein phosphorylation networks are prominent in mediating these fast responses, shaping the cellular response. In the past decade, plant sciences have moved our understanding of these networks further; however, current understanding is largely limited to minutes- and hour-long timescales, and hardly illuminates the (sub-)second timescales at which the initial signaling processes take place. In this Tansley insight, we discuss how quantitative mass spectrometry-based phosphoproteomics can be utilized to study the rapid and dynamic initial steps of protein phosphorylation networks in plants. We highlight rapid responses and show how bioinformatic approaches and the integration of other proteomics approaches can be utilized to deconvolve phosphorylation-based signaling in a data-driven approach. We offer an outlook on how to experimentally address rapid signaling and hope to inspire new approaches in the study of phosphorylation-dependent plant signaling networks.

|
Scooped by
mhryu@live.com
Today, 5:43 PM
|
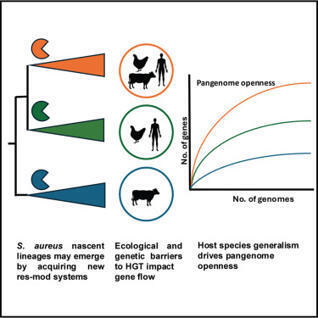
Horizontal gene transfer (HGT) is a major driver of diversity in bacterial populations. However, our understanding of its impact on the evolution of intra-species lineages is limited. The multi-host bacterial pathogen Staphylococcus aureus is differentiated into genetic lineages known as clonal complexes (CC) with variable host and disease tropisms. Here, we demonstrate that CCs exhibit extensive variation in pangenome size, structure, and gene flow, influenced by both genetic and ecological barriers to HGT. Examination of pangenome openness for each CC revealed remarkable variation that correlated strongly with host-species promiscuity. Notably, CCs were defined by horizontally acquired defense systems, and genetic subpopulations have diverged by changes to their type I restriction-modification (R-M) system repertoire, suggesting a role in lineage emergence. Overall, our data indicate a key role of HGT of defense systems in promoting the differentiation of S. aureus into lineages, with host ecology as a major driver of accessory genome variation.

|
Scooped by
mhryu@live.com
Today, 1:26 PM
|
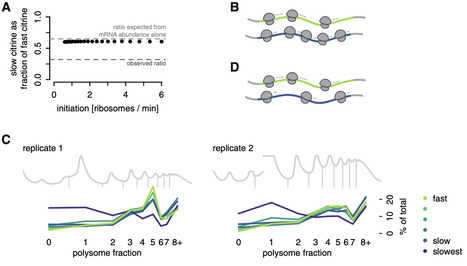
Synonymous codons are decoded at different speeds, but simple models predict that this should not drive protein output: translation initiation, not elongation, should limit the rate of protein production. We showed previously that the output of a series of synonymous fluorescent reporters in yeast spanned a 7-fold range corresponding to translation elongation speed. Here, we show that this effect is not due primarily to the established impact of slow elongation on mRNA stability. Rather, slow elongation further decreases the number of proteins made per mRNA. Our simulations, experiments on fluorescent reporters, and analysis of endogenous protein synthesis in yeast show that translation is limited on non-optimally encoded transcripts. Using a genome-wide CRISPRi screen, we find that impairing initiation attenuates the impact of slow elongation, showing a dynamic balance between rate-limiting steps of protein production. Our results show that codon choice can directly limit protein production across the full range of endogenous codon usage.

|
Scooped by
mhryu@live.com
Today, 1:06 PM
|
Regulation of translation initiation is central to bacterial adaptation, but species-specific mechanisms remain poorly understood. We present high-resolution mapping of translation start sites in Staphylococcus aureus, revealing distinct features of initiation alongside numerous unannotated small ORFs. Our analysis, combined with cryo-EM of a native mRNA-ribosome complex, shows that S. aureus relies on extended, start codon proximal Shine-Dalgarno (SD) interactions, creating specificity against phylogenetically distant bacteria. Several natural S. aureus initiation sites are not correctly decoded by E. coli ribosomes. We identify new and conserved non-canonical start codons, whose regulatory initiation sites contain these characteristic extended SD sequence motifs. Finally, we characterize a novel example of uORF-mediated translational control in S. aureus, demonstrating that translation of a small leader peptide modulates expression of a key biofilm regulator. The described mechanism involves codon rarity, ribosome pausing, and arginine availability, linking nutrient sensing to biofilm formation in this major human pathogen. Kohl et al. combine high-resolution Ribo-seq and cryo electron microscopy to show that the bacterium Staphylococcus aureus uses extended Shine-Dalgarno motifs to initiate translation, which can make start-site decoding incompatible with phylogenetically distant ribosomes.

|
Scooped by
mhryu@live.com
Today, 12:13 PM
|
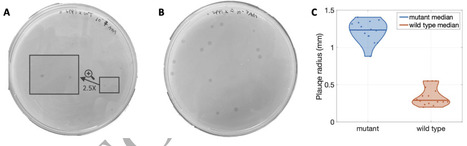
Bacteriophage infect, lyse, and propagate within bacterial populations. However, physiological changes in bacterial cell state can protect against infection even within genetically susceptible populations. One such example is the generation of endospores by Bacillus and its relatives, characterized by a reversible state of reduced metabolic activity that protects cells against stressors including desiccation, energy limitation, antibiotics, and infection by phage. Here we tested how sporulation at the cellular scale impacts phage dynamics at population scales when propagating amongst B. subtilis in spatially structured environments. Plaques resulting from infection and lysis were approximately 3-fold smaller on lawns of spore-forming bacteria vs. non-spore-forming bacteria. Analysis of plaque growth revealed that final plaque size was reduced due to an early termination of expanding phage plaques rather than the reduction of plaque growth speed. Microscopic imaging of the plaques revealed “sporulation rings”, i.e., spores enriched around plaque edges relative to phage-free regions. We developed a series of mathematical models of phage, bacteria, spore, and small molecules that recapitulate plaque dynamics. We show evidence that phage infections trigger the formation of sporulation rings that reduce the productivity of phage infections and halt plaque spread even when resources are available for infection and lysis further away from plaque centers. Moreover, sporulation rings are also enriched in viable virospores, suggesting that although dormancy limits phage infections at population scales in the near-term, viruses may co-opt phage-avoidance strategies to re-emerge over the long-term, opening new avenues to explore the entangled fates of phages and their bacterial hosts.

|
Scooped by
mhryu@live.com
Today, 11:55 AM
|
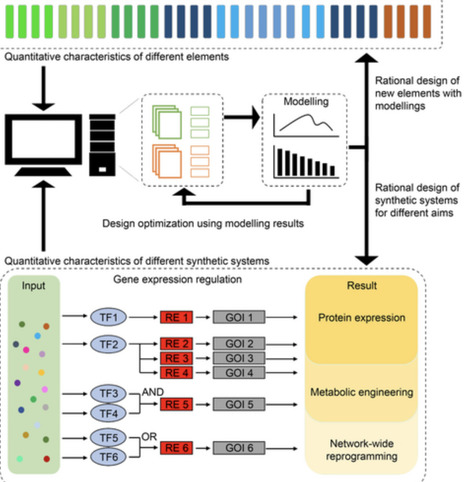
The flourishing plant science promotes the exploding number of data and the expansion of toolkits. Plant synthetic biology is still in its early stages and requires more quantitative and predictable study. Despite the challenges, some pioneering examples have been successfully demonstrated in model plants. As an increasing number of synthetic switches and circuits have been created for plant systems and of synthetic products produced in plant chassis, plant synthetic biology is taking a strong foothold in agriculture and medicine. The ever-exploding data has also promoted the expansion of toolkits in this field. Genetic parts libraries and quantitative characterization approaches have been developed. However, plant synthetic biology is still in its infancy. The considerations for selecting biological parts to design and construct genetic circuits with predictable functions remain desired. In this article, we review the current biotechnological progresses in field of plant synthetic biology. Assembly standardization and quantitative approaches of genetic parts and genetic circuits are discussed. We also highlight the main challenges in the iterative cycles of design-build-test-learn for introducing novel traits into plants. Plant synthetic biology promises to provide important solutions to many issues in agricultural production, human health care, and environmental sustainability. However, tremendous challenges exist in this field. For example, the quantitative characterization of genetic parts is limited; the orthogonality and the transfer functions of circuits are unpredictable; and also, the mathematical modeling-assisted circuits design still needs to improve predictability and reliability. These challenges are expected to be resolved in the near future as interests in this field are intensifying. synbio

|
Scooped by
mhryu@live.com
Today, 11:47 AM
|
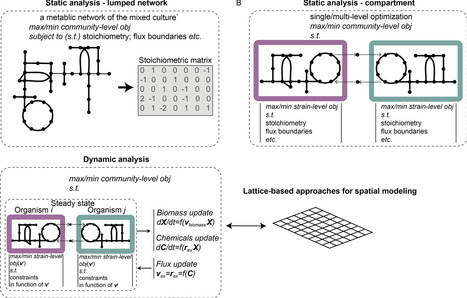
Synthetic microbial communities syncom, with different strains brought together by balancing their nutrition and promoting their interactions, demonstrate great advantages for exploring complex performance of communities and for further biotechnology applications. The potential of such microbial communities has not been explored, due to our limited knowledge of the extremely complex microbial interactions that are involved in designing and controlling effective and stable communities. Genome-scale metabolic models (GEM) have been demonstrated as an effective tool for predicting and guiding the investigation and design of microbial communities, since they can explicitly and efficiently predict the phenotype of organisms from their genotypic data and can be used to explore the molecular mechanisms of microbe-habitats and microbe-microbe interactions. In this work, we reviewed two main categories of GEM-based approaches and three uses related to design of syncom: predicting multi-species interactions, exploring environmental impacts on microbial phenotypes, and optimizing community-level performance. Although at the infancy stage, GEM-based approaches exhibit an increasing scope of applications in designing syncom. Compared to other methods, especially the use of laboratory cultures, GEM-based approaches can greatly decrease the trial-and-error cost of various procedures for designing syncom and improving their functionality, such as identifying community members, determining media composition, evaluating microbial interaction potential or selecting the best community configuration. Future efforts should be made to overcome the limitations of the approaches, ranging from quality control of GEM reconstructions to community-level modeling algorithms, so that more applications of GEMs in studying phenotypes of microbial communities can be expected.

|
Scooped by
mhryu@live.com
Today, 11:36 AM
|
Bacterial colonies, as dynamic ecosystems, display intricate behaviors and organizational structures that profoundly influence their survival and functionality. These communities engage in physiological and social interactions, resulting in remarkable spatial heterogeneity. Recent advancements in technology and modeling have significantly enhanced our comprehension of these phenomena, shedding light on the underlying mechanisms governing bacterial colony development. In this review, we explore the multifaceted aspects of bacterial colonies, emphasizing their physiological intricacies, innovative research tools, and predictive modeling approaches. By integrating diverse perspectives, we aim to deepen our understanding of these microbial communities and pave the way for novel applications in biotechnology, ecology, and medicine.

|
Scooped by
mhryu@live.com
Today, 11:27 AM
|
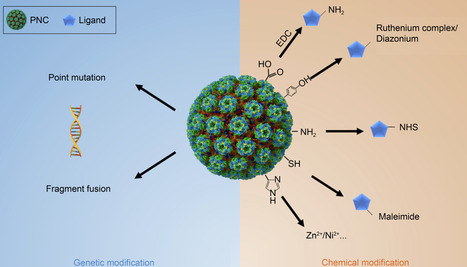
As one of the representative protein materials, protein nanocages (PNCs) are self-assembled supramolecular structures with multiple advantages, such as good monodispersity, biocompatibility, structural addressability, and facile production. Precise quantitative functionalization is essential to the construction of PNCs with designed purposes. With three modifiable interfaces, the interior surface, outer surface, and interfaces between building blocks, PNCs can serve as an ideal platform for precise multi-functionalization studies and applications. This review summarizes the currently available methods for precise quantitative functionalization of PNCs and highlights the significance of precise quantitative control in fabricating PNC-based materials or devices. These methods can be categorized into three groups, genetic, chemical, and combined modification. This review would be constructive for those who work with biosynthetic PNCs in diverse fields.

|
Scooped by
mhryu@live.com
Today, 11:04 AM
|
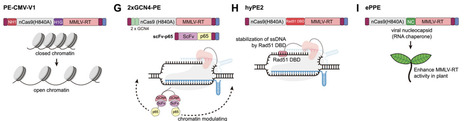
Genetic mutations cause approximately 80% of rare human diseases, highlighting the urgent need for precise genome editing. Since CRISPR-cas9 nucleases were first used for genome editing in 2012, genome editing technologies have rapidly advanced. Base editors, derived from the CRISPR-Cas system, were developed to introduce specific point mutations without requiring DNA double-strand breaks, and subsequently, prime editing (PE) technology was created to enable insertions, deletions, and all types of point mutations. The precision and versatility of PE make it a promising tool for clinical applications. However, PE has potential limitations, including low editing efficiency and limited capacity for large-scale manipulation. To overcome these limitations, research has been continuously conducted to improve PE efficiency and expand its capabilities. Therefore, this review aims to highlight current efforts and future directions for developing and improving PE-related tools.

|
Scooped by
mhryu@live.com
Today, 1:30 AM
|
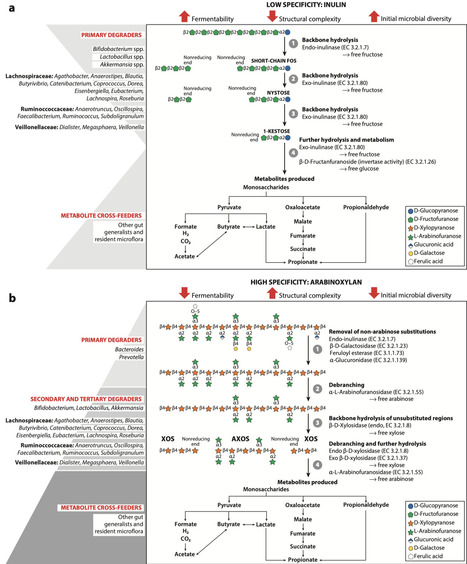
Dietary fibers are crucial in shaping gut microbial composition and functionality. Physical complexity and chemical interactions between fibers and the gut environment lead to diverse and specialized responses that involve entire food webs of gut bacteria; however, there is comparatively less emphasis on understanding ecological dynamics to predict these outcomes. These responses may potentially promote either a broader (less specific) or narrower (more specific) group of gut bacterial taxa, which may vary across individuals. This review examines fiber specificity at the organismal and community levels by exploring mechanistic interactions among dietary fibers and gut bacteria. We discuss the interplay of exogenous and endogenous factors and the structure–function relationships influencing fiber specificity. We establish a mathematical framework to describe specificity in fiber–microbiome interactions based on directionality, magnitude, and stochasticity of fiber–microbiome ecological responses. Finally, we identify research gaps to enhance fiber–microbiota predictions, with implications for strategies aimed at optimizing fiber design.
|
 Your new post is loading...
Your new post is loading...