 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 4:27 PM
|
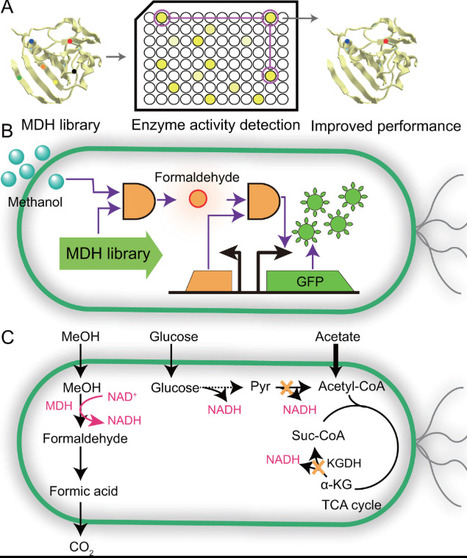
Methanol, a renewable non-food C1 substrate, holds great promise as a feedstock for sustainable biomanufacturing and carbon neutral production. However, its industrial application is hindered by low methanol assimilation efficiency in most microbes. Recent advances in synthetic biology and metabolic engineering have enabled the development of methylotrophic microbial cell factories through strategies including building efficient methanol-utilizing pathways, engineering methanol dehydrogenase for enhanced oxidation efficiency, and optimizing redox balance via cofactor utilization. Additionally, approaches such as mitigating the accumulation of toxic metabolites and adaptive laboratory evolution have been adopted to improve the robustness of synthetic methylotrophs. This review summarizes these innovations and provides a blueprint for rationally designing high-performance microbial platforms to facilitate industrial methanol utilization and advance sustainable development.

|
Scooped by
mhryu@live.com
Today, 1:44 PM
|
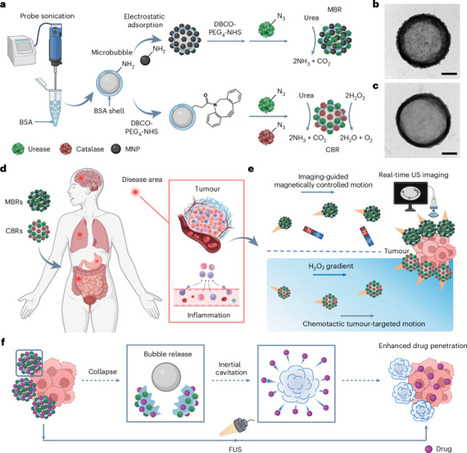
The development of micro- and nano-robots has amplified the demand for intelligent multifunctional machines in biomedical applications, but most microrobotic systems struggle to achieve the attributes needed for those applications. Here we introduce enzymatic microbubble robots that exhibit steerable motion, enhanced biodegradability, high in vivo imaging contrast, and effective targeting and penetration of disease sites. These microrobots feature natural protein shells modified with urease to decompose bioavailable urea for autonomous propulsion, whereas an internal microbubble serves as an ultrasound imaging contrast agent for deep tissue imaging and navigation. Magnetic nanoparticle integration enables imaging-guided magnetically controlled motion and catalase functionalization facilitates chemotactic movement towards hydrogen peroxide gradients, directing robots to tumour sites. Focused ultrasound triggers robot shell collapse and inertial cavitation of the released microbubbles, creating mechanical forces that enhance therapeutic payload penetration. In vivo studies validate the tumour-targeting and therapeutic efficacy of these robots, demonstrating enhanced antitumour effects. This multifunctional microbubble robotic platform has the potential to transform medical interventions and precision therapies. Biodegradable enzymatic microbubble robots self-propel in urea, are magnetically or chemotactically guided, provide ultrasound imaging and enhance intratumoural drug delivery with focused ultrasound.

|
Scooped by
mhryu@live.com
Today, 12:59 PM
|
Biological nitrogen fixation (BNF) and photosynthetic carbon fixation underpin food production and climate mitigation, yet natural systems are constrained by oxygen sensitivity, high energy demand, and inefficient catalysts. This review synthesizes advances that recast these processes as engineering targets and proposes a conceptual roadmap that bridges synthetic symbioses with the synthetic biology of enzymes and pathways. For BNF, progress spans cross-kingdom strategies—from refactoring nif gene sets and targeting nitrogenase assembly to eukaryotic organelles, to engineering plant-associated diazotrophs, rhizosphere control circuits, and emerging nodule-like microenvironments. For carbon assimilation, new-to-nature CO2-fixation modules and photorespiratory bypasses illustrate how pathway redesign and alternative carboxylases can circumvent key Calvin–Benson–Bassham limitations, and expanding photosynthetic light capture offers additional leverage. Across these domains, we extract common design principles: (i) nitrogenase output is increasingly governed by carbon/energy supply and electron delivery as much as by oxygen protection; (ii) robust function requires compartment-aware enzyme–chassis coordination, substrate channeling, and dynamic regulation using sensors and control circuits; and (iii) scalable implementation may benefit from distributing metabolic labor across engineered consortia rather than forcing all functions into a single host. We discuss enabling technologies—including AI-guided protein design and directed evolution, cell-free prototyping, chassis toolkits, and materials/bioelectrochemical interfaces—that can accelerate design–build–test–learn cycles and reduce barriers to deployment. Together, these insights define a path toward integrated nitrogen and carbon fixation systems for low-emission agriculture and biomanufacturing.

|
Scooped by
mhryu@live.com
Today, 12:48 PM
|
Bacterial contractile injection systems (CISs) are multiprotein complexes that facilitate the bacterial response to environmental factors or interactions with other organisms. Multiple novel CISs have been characterised in laboratory bacterial cultures recently; however, studying CISs in the context of the native microbial community remains challenging. Here, we present an approach to characterise a bioinformatically predicted CIS by directly analysing bacterial cells from their natural environment. Using cryo-focused ion beam milling and cryo-electron tomography (cryoET) imaging, guided by 16S rRNA gene amplicon sequencing, we discovered that thermophilic Chloroflexota bacteria produce intracellular CIS particles in a natural hot spring microbial mat. We then found a niche-specific production of CIS in the structured microbial community using an approach combining metagenomics, proteomics, and immunogold staining. Bioinformatic analysis and imaging revealed CISs in other extremophilic Chloroflexota and Deinococcota. This Chloroflexota/Deinococcota CIS lineage shows phylogenetic and structural similarity to previously described cytoplasmic CIS from Streptomyces and probably shares the same cytoplasmic mode of action. Our integrated environmental cryoET approach is suitable for discovering and characterizing novel macromolecular complexes in environmental samples.

|
Scooped by
mhryu@live.com
Today, 12:31 PM
|
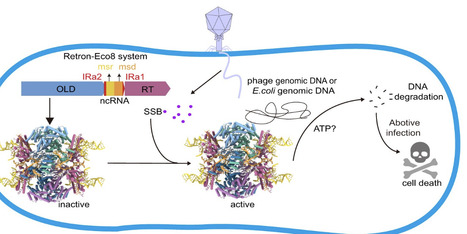
Retrons represent a novel class of bacterial defense systems that employ reverse transcriptase (RT), noncoding RNA, and effector proteins to counteract phage infections. In this study, we elucidate the molecular mechanism of a retron system, Retron–Eco8. Biochemical experiments reveal that the Retron–Eco8 holocomplex, rather than the effector alone, exhibits double-stranded DNA cleavage activity, triggering abortive infection and therefore effectively halting phage propagation. Cryo-electron microscopy (cryo-EM) analysis reveals a supramolecular assembly comprising four RT subunits, four multicopy single-stranded DNA molecules, and four overcoming lysogenization defect (OLD) nucleases—a configuration critical for antiphage defense. Notably, we examine the activation of Retron–Eco8 by diverse single-stranded DNA-binding (SSB) proteins, and phylogenetic analysis of these SSB proteins elucidates the phage resistance specificity. Collectively, our findings delineate the structural architecture of the Retron–Eco8 defense complex and provide mechanistic insights into retron-mediated bacterial immunity.

|
Scooped by
mhryu@live.com
Today, 10:44 AM
|
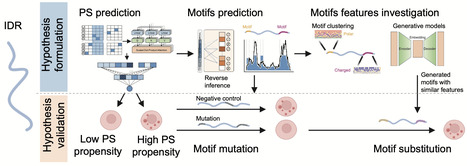
Intrinsically disordered regions (IDRs) in proteins drive phase separation (PS) to form biomolecular condensates, which organize cellular matter. While IDRs are recognized as critical drivers of PS, the systematic identification of sequence motifs governing this phenomenon and their compositional determinants remain a key challenge. Here we develop PhaSeMotif, a deep learning framework for interpretable and precise predictions of essential phase-separating motifs within IDRs. We experimentally validate PhaSeMotif, demonstrating that mutations of predicted motifs significantly reduce or eliminate the PS capabilities of IDRs. The identified motifs possess diverse amino acid compositional features that are critical for determining PS propensities and condensate partitioning. Furthermore, PhaSeMotif integrates generative models to create validation-ready motifs that preserve these critical compositional features, empowering direct experimental verification and deeper mechanistic investigation of PS-driving IDR motifs. Overall, by combining motifs prediction, generation, and validation, PhaSeMotif provides an open-access toolkit to facilitate more efficient IDR motifs investigation and provides insights into the molecular determinants of PS. Deciphering the rules of protein phase separation remains a challenge. Here, the authors develop PhaSeMotif, an interpretable and generative deep learning framework that identifies essential sequence motifs and designs functional synthetic variants.

|
Scooped by
mhryu@live.com
Today, 10:23 AM
|
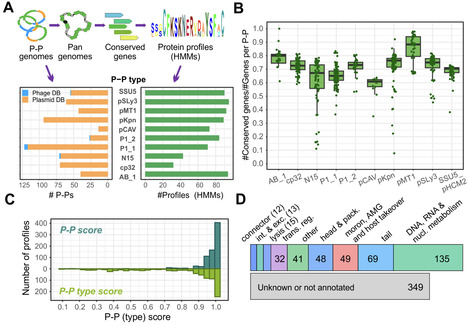
Phage-plasmids (P-Ps) are temperate phages that replicate as plasmids during lysogeny. Despite their high diversity, they carry genes similar to phages and plasmids. This leads to gene exchanges and to the formation of hybrid or defective elements, which limits accurate detection of P-Ps. To address this challenge, we developed tyPPing, an easy-to-use method that efficiently detects and types P-Ps with high accuracy. It searches for distinct frequencies and sets of conserved proteins to separate P-Ps from plasmids and phages. tyPPing’s strength comes from both its precise predictions and its ability to systematically type P-Ps, including the assignment of confidence levels. We tested tyPPing on several databases and a collection of incomplete (draft) genomes. While predictions rely on the quality of assemblies, we detected high-quality P-Ps and experimentally proved them to be functional. Compared to other classification methods, tyPPing is designed to detect distinct P-P types and surpasses other tools in terms of sensitivity and scalability. P-Ps are highly diverse, making the systematic identification of new types a difficult task. By combining tyPPing with other tools, however, we show a valuable foundation for addressing this challenge. How to use tyPPing and other approaches is documented in our GitHub repository: github.com/EpfeiferNutri/Phage-plasmids/.

|
Scooped by
mhryu@live.com
Today, 9:27 AM
|
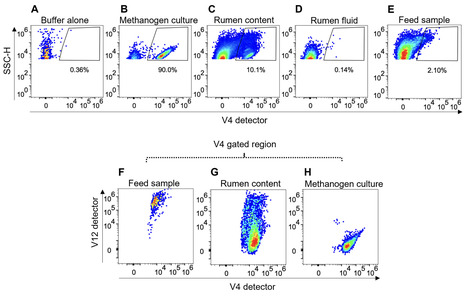
Methanogenic archaea that reside in the rumen of sheep, cattle, and other ruminants generate 16% of global emissions of methane, a potent greenhouse gas. The majority of rumen methanogens belong to species that display readily observable autofluorescence due to their intracellular co-factor, F420. We developed a spectral flow cytometry method to directly quantify autofluorescent methanogens in the complex environment of the rumen. Rumen samples contain feed particles with natural autofluorescence signatures that overlapped those of F420-containing methanogens. Spectral unmixing using natural autofluorescence signatures allowed us to distinguish methanogens from other autofluorescent particles and to quantify both cultured methanogens in buffer and native methanogens in rumen content samples over a concentration range from 4 × 104 to 4 × 107 cells/mL. The methanogen signal was absent in microbial cultures known to lack F420 and in rumen content samples treated with sodium borohydride (NaBH4), which reduces F420 fluorescence. We showed a strong relationship between the number of autofluorescent methanogens in rumen content samples and methane yields in cattle and sheep treated with a methanogen inhibitor. We also assessed the impact of sample fixation on the spectral profiles of methanogen cells and showed that rumen samples stored at 4°C for up to 3 days remain suitable for enumeration. Our data thus demonstrate a new spectral flow cytometry method that can be used for rapid quantification of autofluorescent methanogens in rumen content samples.

|
Scooped by
mhryu@live.com
Today, 1:48 AM
|
Trichoderma atroviride is well-known biocontrol fungus that plays a crucial role in controlling plant fungal diseases. In this study, the Serine Protease (T. atSp1) gene of T. atroviride was selected as the target gene to investigate the effects of Agrobacterium tumefaciens concentration, conidial concentration, mixing ratio of conidia and Agrobacterium cells, and induction time on transformation efficiency. The optimal knockout system was achieved under conditions that the density of A. tumefaciens was OD600 = 0.5, the concentration of conidia suspension was 106 conidia/mL, the mixture ratio of conidia suspension and A. tumefaciens AGL-1 was 1:1, and the induction time was 0.5 h. The transformation efficiency reached 28.33 to 61.67 transformants per 106 conidia under the optimized conditions. The ΔT. atSp1 was successfully validated by PCR analysis. Additionally, two genes of T. atEDG1 and T. atchi18 were also knocked out and verified, further demonstrating the robustness of this ATMT system. This study provides a stable and efficient genetic manipulation protocol for T. atroviride, facilitating further to understand genes function and biocontrol mechanisms.

|
Scooped by
mhryu@live.com
Today, 1:41 AM
|
Covalent bond–forming peptide tagging systems have emerged as powerful and versatile tools across a broad spectrum of biological and biotechnological applications. This review systematically summarizes the origins, molecular mechanisms of intramolecular covalent bond formation, major classes, and design strategies of peptide tagging systems. Based on their underlying chemistry, current systems are primarily categorized into isopeptide-bond-based and ester-bond-based platforms, both of which have demonstrated prominent utility in protein cyclization as well as in vivo and in vitro multi-enzyme assembly. Beyond these applications, isopeptide-bond-forming systems have been widely adopted as robust purification tags, whereas ester-bond-based systems offer unique opportunities for pH-responsive modulation of enzyme activity. Collectively, peptide tagging systems based on either isopeptide or ester bond formation represent an expanding and highly efficient toolkit for biotechnology. Continued advances in their design and application are expected to further broaden their functional scope and provide innovative solutions for future developments in protein engineering and related fields.

|
Scooped by
mhryu@live.com
Today, 1:20 AM
|
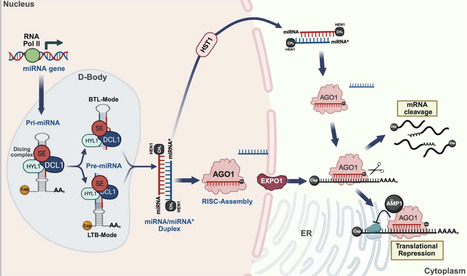
Plant branching, encompassing both vegetative and reproductive forms, is a complex and crucial process that shapes overall architecture and determines crop yield and biomass. MicroRNAs (miRNAs) have emerged as master regulators in fine-tuning the intricate genetic and hormonal networks that govern plant branching. This review systematically synthesises recent advances in understanding how miRNA-target gene modules regulate essential pathways to orchestrate the branching patterns. We highlight a central insight that specific miRNA families form hierarchical, stage-specific networks that facilitate the independent optimization of vegetative and reproductive branching. Furthermore, we explore the potential applications of miRNA manipulation in optimizing branching architecture to improve crop yield. By critically evaluating strategies such as artificial miRNAs, target mimics and CRISPR/Cas9 genome editing, we discuss how modulating miRNA networks can engineer ideal plant architecture. Finally, we provide a forward-looking perspective on overcoming challenges in miRNA-based crop improvement, emphasising the integration of single-cell omics and epigenetic insights to achieve precise genetic modifications. This review underscores the transformative potential of miRNAs in designing future crops for enhanced productivity.

|
Scooped by
mhryu@live.com
Today, 1:14 AM
|
All pathogens must sense that they have arrived at their host. This is a necessary part of infection in order to effect the changes in pathogen biology required to progress through their life cycle. How the information that they have arrived is transmitted, and what molecules/media convey the information, is poorly understood. Here, we review recent literature and provide speculation as to how this might happen, by analogy to the five human senses. Our criteria center on natural selection: we consider host-derived signals—in the broadest sense—to be those that carry some information and that can be detected by the pathogen, in principle. For each, we identify supporting literature and speculate on areas of possible expansion. We conclude, on the one hand, that there is a diversity of understudied but compelling signals, but, on the other hand, that not all signals are equal. The magnitude of the response is likely a function of the fidelity of the signal/detection. Although knowledge is currently incomplete, the prospect of understanding perception of arrival at the host may allow us to perturb pathogen perception of the host and thereby thwart this early and fundamental step in pathogen development.

|
Scooped by
mhryu@live.com
Today, 1:09 AM
|
RNA structure prediction remains one of the most challenging problems in computational biology, with significant implications for understanding gene regulation, drug design, and synthetic biology. While deep learning has revolutionized protein structure prediction, RNA presents unique challenges including limited training data, complex noncanonical interactions, and conformational flexibility. This review examines the evolution from traditional physics-based methods to current deep learning approaches for RNA secondary and tertiary structure prediction. After briefly exploring traditional methods, like Direct Coupling Analysis and physics-based simulations, we systematically review three deep learning paradigms: language model–based methods, end-to-end structure predictors, and geometry-distance prediction approaches. Furthermore, we identify critical future research directions focusing on advanced tokenization strategies to address data scarcity and explainable artificial intelligence techniques to improve model interpretability. Despite significant progress, achieving transformative performance requires continued methodological innovation, specifically designed for RNA’s unique characteristics, and a substantial expansion of high-quality structural datasets.
|

|
Scooped by
mhryu@live.com
Today, 3:50 PM
|
De novo protein design expands the functional protein universe beyond natural evolution, offering vast therapeutic and industrial potential. Monte Carlo sampling in protein design is under-explored due to the typically long simulation times required or prohibitive time requirements of current structure prediction oracles. Here we make use of a 20-letter structure-inspired alphabet derived from protein language model embeddings to score random mutagenesis-based Metropolis sampling of amino acid sequences. This facilitates fast template-guided and unconditional design, generating sequences that satisfy in silico designability criteria without known homologues. Ultimately, this unlocks a new path to fast and de novo protein design.

|
Scooped by
mhryu@live.com
Today, 1:07 PM
|
Enzymes catalyze diverse chemical transformations and offer a sustainable approach to both breaking and making chemical bonds. However, finding an enzyme capable of performing a specific chemical reaction remains a challenge. We developed a new framework, Enzyme-toolkit (Enzyme-tk), that integrates 23 open-source tools to enable the discovery of enzymes that have activity toward a specific target reaction. Additionally, we introduce two new methods to facilitate enzyme discovery: (1) Func-e, an ML tool that searches large databases for enzymes that potentially catalyze a specific chemical transformation and (2) Oligopoolio, a gene assembly approach that reduces the cost of accessing protein sequences and thus the barrier to their experimental validation. We applied Enzyme-tk to find enzymes for chemical degradation of two man-made pollutants, di-(2-ethylhexyl) phthalate (DEHP) and triphenyl phosphate (TPP). We demonstrate that new, previously unannotated enzymes with favorable characteristics, such as high thermostability, can be identified using Enzyme-tk for reactions that are dissimilar to the training set.

|
Scooped by
mhryu@live.com
Today, 12:55 PM
|
Thiamine (Vitamin B1), an essential water-soluble vitamin, is composed of a pyrimidine and a thiazole ring. Owing to its functional roles as a coenzyme and its anti-inflammatory and antioxidant properties, it plays a critical role in disease prevention and therapeutic interventions. Currently, industrial production of thiamine relies primarily on chemical synthesis-a process that generates significant amounts of hazardous waste and byproducts. In contrast, microbial biosynthesis represents a more sustainable and environmentally friendly alternative. This review first outlines thiamine metabolism in microorganisms, highlighting ThiC as the key rate-limiting enzyme in its biosynthesis. It then summarizes potential strategies for improving thiamine biomanufacturing, and proposes that optimizing metabolic flux together with energy and cofactor balance at critical nodal points is essential for overcoming current yield limitations. Finally, to overcome specific bottlenecks in thiamine biosynthesis, such as precursor transport and pathway optimization, we propose that transport engineering and gene mining represent promising strategies complementary to recent advances in enzyme-directed evolution and metabolic engineering.

|
Scooped by
mhryu@live.com
Today, 12:44 PM
|
Rhodospirillum rubrum owns a dynamic poly(3-hydroxybutyrate) (PHB) cycle: During growth PHB is accumulated and subsequently degraded under carbon starvation. Interestingly, this cycle is typically found for acetate grown R. rubrum but not for fructose grown cells, where no PHB accumulation has been observed. This study aimed to determine whether expression of PHB cycle genes correlates with the phases of PHB accumulation and degradation on acetate in comparison to absence of PHB synthesis during growth on fructose and ΔphaC1ΔphaC2 mutant unable to polymerize PHB on acetate. Surprisingly, transcriptomic analyses of the wild-type strain demonstrated that PHB cycle genes were not only expressed during growth on acetate but also for growth on fructose, regardless of PHB content. Substrate-specific expression patterns were identified: The PHB depolymerase gene phaZ1 was predominantly expressed on acetate, while phaZ2 and the depolymerase regulator apdA were upregulated on fructose. Interestingly, phaC3 and phaZ3 showed distinct expression patterns compared with other PHB cycle genes, particularly in mutant strains. Despite the absence of PHB granules in the ΔphaC1ΔphaC2 strain, several PHB cycle genes remained expressed, and volatile fatty acid assimilation pathways were transcriptionally impacted. These findings highlight the complexity of the PHB cycle and suggest that PHB participates in other physiological processes, such as substrate assimilation, potentially via regulatory actions of PHB granule bound regulator PhaR.

|
Scooped by
mhryu@live.com
Today, 10:47 AM
|
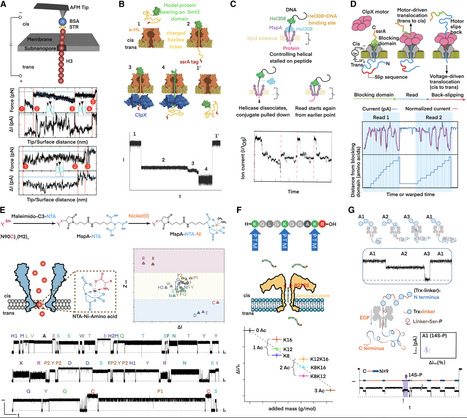
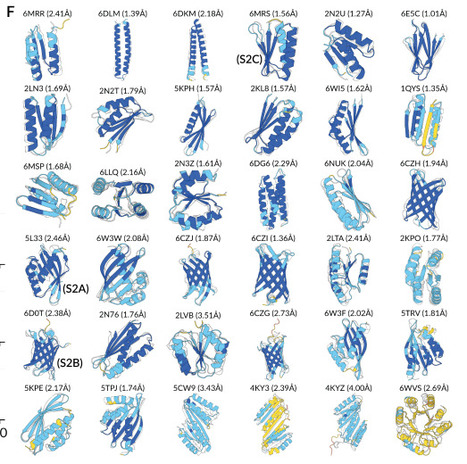
The first nanopore-based sequencer was launched in 2014, and subsequently, nanopore played an irreplaceable role in disclosing the first complete, gapless sequence of a human genome in 2022 due to its megabase-scale read lengths. However, a striking revelation from DNA sequencing is that over 95% of human DNA does not specify a protein, which means tremendous proteomic information cannot be predicted from the genome. Therefore, nanopore researchers have been leaning increasing attention to the proteome. Nowadays, nanopores have demonstrated unprecedented performance in discriminating individual proteinogenic amino acids with chemical modifications. Meanwhile, diverse strategies for full-length proteins to translocate through nanopores have been developed. Undoubtedly, nanopore will sooner or later facilitate de novo protein sequencing. This nanopore review begins with DNA sequencing and elaborates on up-to-date technical breakthroughs in protein sequencing and other proteomics approaches. Overall, nanopore technology is conducive to discovering the proteome diversity and revealing the pathogenesis mechanism.

|
Scooped by
mhryu@live.com
Today, 10:36 AM
|
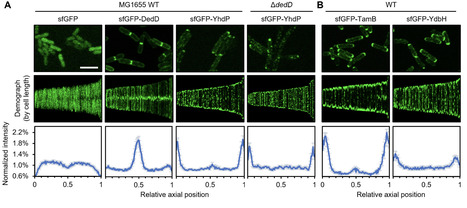
The Gram-negative bacterial cell envelope comprises an outer membrane (OM) with an asymmetric arrangement of lipopolysaccharides and phospholipids (PLs), protecting them from both physical and chemical threats. To build the OM, PLs must be transported across the cell envelope; this process has remained elusive until recently, where three collectively essential AsmA-superfamily proteins—YhdP, TamB, and YdbH—are proposed to function as anterograde PL transporters in E. coli. Here, we identify the cell wall-binding protein DedD as a novel interacting partner of YhdP and discover that all three AsmA-superfamily proteins are recruited to and strongly enriched at the cell poles. Our observation raises the possibility that anterograde PL transport could be spatially restricted to the cell poles and highlights the importance of understanding the spatial-temporal regulation of OM biogenesis in coordination with cell growth and division.

|
Scooped by
mhryu@live.com
Today, 9:59 AM
|
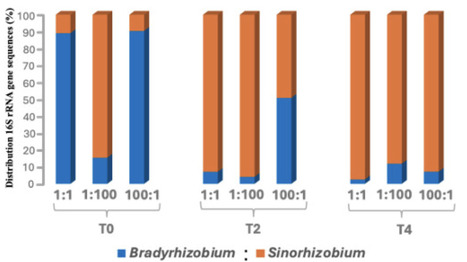
Soybean is frequently nodulated by species from the Bradyrhizobium (BR) and/or Sinorhizobium (SR) genera. Several factors, such as soil pH, host genotype, geographic location, and other environmental variables, are reported to influence the preferential selection between BR and SR species within soybean root nodules. However, it remains unclear whether the age of the host plant at the time of inoculation affects preferential rhizobial selection. To investigate this, we inoculated soybean plants with different cell densities of BR and SR strains at three time points: at sowing (T₀), 2 weeks after germination (T₂), and 4 weeks after germination (T₄). We used 16S rRNA gene amplicon sequencing of root nodules and rhizosphere samples to assess the relative abundance of BR and SR in nodules and rhizosphere. We observed a clear shift in nodule occupancy that favored BR at the time of seed sowing (T₀) but increasingly favored SR when plants were inoculated at T₂ and T₄ stages. Specifically, at T₄, SR dominated in nodules across all treatments, representing 88%–99% of total sequences, regardless of applied inoculum ratio. In contrast, a similar number of sequences for both strains was detected in the rhizosphere at the time of the final harvest. These results highlight host age as an important ecological driver in legume–rhizobium interactions and suggest that inoculation time strongly influences microsymbiont selection. This information is important in understanding rhizobial competition and optimizing the timing of inoculation for soybeans.

|
Scooped by
mhryu@live.com
Today, 1:57 AM
|
Echinocandin B (ECB), a cyclic lipohexapeptide for synthesizing antifungal drugs, is produced by the nonribosomal peptide synthetase gene cluster in Aspergillus nidulans. However, industrial production remains limited by the inefficiency of production capacity, primarily due to the complexity of the biosynthetic pathway and the absence of multi-gene regulatory tools in filamentous fungi. Here, we established an orthogonal Cre-lox-based platform enabling single-site insertion of up to 30 kb and simultaneous dual-site integration of 10 kb DNA fragments in A. nidulans. Through precursor supplementation and targeted gene overexpression, we identified key enzymatic bottlenecks in the precursor biosynthetic pathway, including the oxygenases AniF, AniK, AniG, and the acyl-AMP ligase AniI. Combinatorial overexpression of these genes acted synergistically to increase ECB titers. We further addressed bottlenecks in natural amino acid biosynthesis by overexpressing feedback-resistant mutants of Hom3 (L-Thr pathway) and LeuC (L-Leu pathway). Additionally, we uncovered a temperature-dependent regulation mechanism whereby low temperature (25 °C) concurrently upregulates both the ECB biosynthetic gene cluster and odeA gene, encoding Δ12-oleic acid desaturase, thereby increasing linoleic acid availability for ECB production. Leveraging our multisite DNA-integration platform to rewire expression of these key genes, we increased ECB production to 3.5 ± 0.2 g/L in a 5-L fed-batch bioreactor, a 2.3-fold improvement that represents the highest titer reported in the literature to date. Our orthogonal dual-site integration strategy and the systematic optimization approach provide a valuable framework for metabolic engineering of complex natural products in filamentous fungi.

|
Scooped by
mhryu@live.com
Today, 1:42 AM
|
Recent advances in protein structure prediction have created high-confidence candidate structures for nearly every known protein-coding gene. At the same time, many software packages have been created to visualize protein structures, protein multiple sequence alignments (MSAs), and protein annotations. However, few software tools can highlight the direct relationship between nucleotide variation of protein-coding genes in genome space and the evolutionary and structural context of that variation in protein space. To help address these needs, we created a suite of robust and reusable JavaScript components to show protein structures, MSAs, phylogenies, and their relationship to protein-coding gene regions using the JBrowse 2 genome browser. This software allows users to interface with web services such as AlphaFoldDB and Foldseek to access pre-computed structures, or to upload protein structures from sources such as ColabFold or PDB. Our resources are available at https://github.com/GMOD/proteinbrowser.

|
Scooped by
mhryu@live.com
Today, 1:38 AM
|
Methanol is a highly promising feedstock for biomanufacturing owing to its broad availability, low cost, and high energy density. Methylotrophic fermentations have been exploited to produce diverse fuels, chemicals, and materials. However, although such processes have been practiced for decades, their applications have been constrained by low methanol assimilation efficiency, insufficient cellular energy and reducing equivalents supply, the cytotoxicity of methanol and its intermediates, and inadequate robustness of chassis strains. In this review, progress is synthesized along four pillars for constructing high-performance methanol bio-converting cell factories: methanol assimilation pathways, energy-supply strategies, tolerance-enhancement approaches, and metabolic engineering for chemical synthesis, with the aim of informing the rational design and construction of efficient methanol bio-converting cell factories.

|
Scooped by
mhryu@live.com
Today, 1:17 AM
|
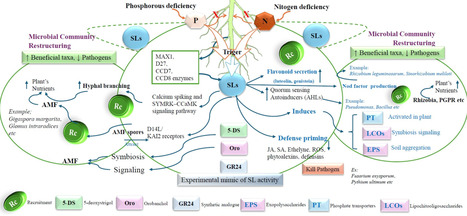
Strigolactones (SLs) are phytohormones derived from carotenoids that influence various aspects of plant growth, development, and the ability of plants to respond to environmental changes and microbial interactions. Initially categorized as shoot branching inhibitors, SLs are now recognized as crucial rhizospheric signaling molecules that govern nutrient availability, hormonal control, and microbial interactions. Despite significant progress in SL biology, a cohesive synthesis connecting SL molecular signaling, rhizosphere communication, and stress tolerance remains fragmented, hindering their practical use in sustainable agriculture. A more comprehensive understanding of their synthesis process (D27-CCD7/8-MAX1-CLA cascade), their perception (D14-MAX2-SMXL module), and the impact of SMXL7 on chromatin has revealed significant implications on physiology. To enhance plant development under stress conditions, SLs drive auxin transport, regulate ABA-dependent stress signaling, influence the antagonistic effects of cytokinins, and coordinate gibberellin activity with the circadian rhythm. SLs augment arbuscular mycorrhizal colonization, stimulate nodulation, and attract plant growth-promoting rhizobacteria through chemotactic and metabolic interactions. Using GR24 and SL-conjugated nanomaterials enhances plant resistance to drought, salt, and metal stress. Modifying SL-transporters with CRISPR improves SL signaling and fosters beneficial symbiotic associations. The study is crucial because it underscores the importance of SLs in recruiting beneficial microorganisms and facilitating microbial-hormonal interactions. This review proposes a cohesive conceptual framework that integrates receptor specificity, rhizospheric sensing, and microbial response, beyond mere descriptive synthesis. It sets distinct research targets, such as receptor-specific SL-analogues, in situ sensing techniques, and tailored SL-responsive microbial consortia, to make biostimulation more precise and assist crops in withstanding climatic stress more effectively.

|
Scooped by
mhryu@live.com
Today, 1:11 AM
|
The pressing challenges posed by climate change and the depletion of traditional energy sources have intensified the search for alternative energy-harvesting technologies. Plant-microbial fuel cells (PMFCs) have emerged as a promising solution. Although they are not yet energetically competitive, their potential application in low-power devices as a battery replacement has been widely explored. PMFCs operate by integrating living plants with microbial fuel cells to generate electricity in situ through the metabolic activity of electroactive microorganisms (EAMs) in the rhizosphere. These microbes degrade root exudates and play a central role in PMFC performance and long-term stability. In this review, we selected 21 studies that examined bacterial and archaeal communities in PMFCs, comparing their microbial composition and resulting electricity outputs. We highlight how differences in plant species, system configurations, and environmental conditions influence the structure and function of microbial communities. We also discuss the methods used for microbial community assessment and address the persistent lack of standardization across studies, which limits comparability. Finally, we outline future research directions aimed at optimising PMFC performance, including the search for electroactivity biomarkers, the potential of genetic engineering and nanomaterials, and the largely unexplored electroactive potential of eukaryotes in these systems. This review advances the existing literature by incorporating recent findings and offering a renewed perspective on PMFC systems.
|
 Your new post is loading...
Your new post is loading...




















2st, nanopore review