 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:07 AM
|
Researchers have published the recipe for an artificial-intelligence model that reviews the scientific literature better than some major large language models (LLMs) are able to, and gets the citations correct as often as human experts do. OpenScholar — which combines a language modelwith a database of 45 million open-access articles — links the information it sources directly back to the literature, to stop the system from making up or ‘hallucinating’ citations. Several commercial AI-based literature-review tools already exist that use similar techniques, but few have been released as open source, says Akari Asai, an AI researcher at Carnegie Mellon University in Pittsburgh, Pennsylvania, and a co-author of the work, published in Nature on 4 February1. Being open source means that researchers can not only try OpenScholar for free in an online demonstration, but also deploy it on their own machine and use the method in the paper to boost the literature-review skills of any LLM, says Asai. In the 14 months since OpenScholar was first published in the arXiv repository2, AI firms such as OpenAI have used similar methods to tack ‘deep research’ tools onto their commercial LLMs, which has greatly improved their accuracy. OpenScholar also outperformed GPT-4o, as well as rival tools such as PaperQA2

|
Scooped by
mhryu@live.com
February 4, 11:37 PM
|
Organofluorine compounds are of considerable concern due to their environmental persistence and human health effects. Their persistence stems from an inability of native microorganisms to metabolize them. Polyfluorinated compounds are unnatural, offer little nutritional benefit to microorganisms, and their breakdown releases toxic fluoride anion. Yet, the biological breakdown of dispersed polyfluorinated compounds would offer a compelling strategy for environmental remediation. In this context, molecular biotechnology is being pursued. The first required tools are methods for monitoring biodefluorination, often determining fluoride release by different methods. The use of those methods revealed fluoride toxicity as a major selection against biodefluorination. Despite that, microbial enzymes catalyzing defluorination of monofluorinated compounds are known and provide templates for bioengineering systems to handle polyfluorinated compounds. Whereas biodegradation often evolves in nature, effective biodegradation of perfluorinated chemicals may require laboratory evolution and engineering.

|
Scooped by
mhryu@live.com
February 4, 10:55 PM
|
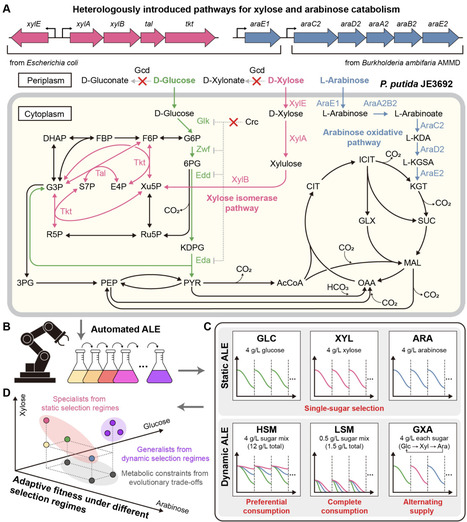
Efficient co-utilization of hexose and pentose sugars from lignocellulose is essential for microbial production of bio-based chemicals, yet engineered non-native catabolic pathways can be suboptimal and evolutionarily unstable in complex resource environments. We used a Pseudomonas putida strain, previously engineered to catabolize xylose and arabinose to examine how resource abundance, temporal availability, and sub-culturing criteria shape evolutionary outcomes. Using an automated adaptive laboratory evolution (ALE) platform, we evolved the strain under static conditions with single selection pressures and dynamic regimes that imposed selection pressures on multiple sugars. These environments drove divergence between catabolic specialists and generalists. While selection regimes with weak or absent selection for xylose frequently resulted in loss of xylose catabolism, evolution under carbon-limited, mixed-sugar environments promoted stable retention and coordinated optimization of multiple catabolic pathways, increasing total sugar consumption in mixed-sugar conditions. Genomic, proteomic, and biochemical analyses showed that evolutionary stability was determined by pathway-specific fitness costs, leading to either pathway loss or cost-reducing refinement, depending on selection strength. An isolated generalist clone also exhibited improved indigoidine production from mixed sugars when compared to the parental strain. Together, these findings link resource dynamics to fitness landscapes that determine catabolic specialization, generalization, evolutionary trade-offs, and applicability to bioconversion.

|
Scooped by
mhryu@live.com
February 4, 10:39 PM
|
An unresolved challenge in the field of computational protein design is to create proteins that bind non-protein partners, e.g. DNA, RNA, and small molecules. Most machine learning (ML) algorithms for protein design can only work with systems composed entirely of amino acids, and therefore cannot be directly applied to this task. The few algorithms that accommodate non-proteins still represent amino acids differently than other molecules, and therefore cannot easily recognize the similarity between a sidechain and a small molecule that share a functional group. We introduce a new method, called AtomPaint, that avoids these limitations by employing a fully-atomic representation of protein structure. Starting from a model of a desired binding interaction, our method proceeds by (i) converting that model to a 3D image, (ii) masking out the parts of that image that need to be redesigned, (iii) using a diffusion model to inpaint the masked voxels, then (iv) using a classification model to identify the amino acids in the inpainted image. Both models are SE(3)-equivariant ResNets, and were trained on a dataset of structures from the Protein Data Bank (PDB) curated to emphasize protein/non-protein interactions. In a sequence recovery benchmark, AtomPaint performed better than random guessing, suggesting that it understands some aspects of molecular structure. We discuss possible avenues of improvement, in the hopes that the advantages of our novel image-based approach can be fully realized.

|
Scooped by
mhryu@live.com
February 4, 2:38 PM
|
The human gastrointestinal tract is home to a diverse community of microorganisms from all domains of life, collectively referred to as the gut microbiome. While gut bacteria have been studied extensively in relation to human host health and physiology, other constituents remain underexplored. This includes the gut virome, the collection of bacteriophages, eukaryotic viruses, and other mobile genetic elements present in the intestine. Like gut bacteria, the gut virome has been causatively linked to human health and disease. However, the gut virome is substantially more difficult to characterize, given its high diversity and complexity, as well as multiple challenges related to in vitro cultivation and in silico detection and annotation. In this mini-review, we describe various methodologies for examining the gut virome using both culture-dependent and culture-independent tools. We highlight in vitro and in vivo approaches to cultivate viruses and characterize viral-bacterial host dynamics, as well as high-throughput screens to interrogate these relationships. We also outline a general workflow for identifying and characterizing uncultivated viral genomes from fecal metagenomes, along with several key considerations throughout the process. More broadly, we aim to highlight the opportunities to synergize and streamline wet- and dry-lab techniques to robustly and comprehensively interrogate the human gut virome.

|
Scooped by
mhryu@live.com
February 4, 2:02 PM
|
Methanotrophs are of fundamental importance in the global methane cycle and hold promise for the bioconversion of methane into valuable products. This study elucidates the impact of alternating aerobic–anoxic conditions on metabolism of the two phylogenetically distinct groups of methanotrophs: Methylomonas koyamae (type I, γ-proteobacteria) and Methylocystis bryophila (type II, α-proteobacteria). Using batch cultivations with single and repeated oxygen pulses, we demonstrated that cyclic aerobic–anoxic alternation significantly increased the secretion of organic compounds. Notably, acetate secretion by the type II methanotroph M. bryophila was observed for the first time. Genome-scale metabolic modeling and flux balance analysis revealed that acetate secretion serves as an auxiliary ATP-generating pathway for M. bryophila under oxygen limitation. We further uncovered a dynamic mechanism: aerobic phases trigger methane uptake and build up intracellular organic carbon pools, while anoxic phases redirect these pools toward acetate and/or hydrogen release to sustain survival under oxygen limitation. This cyclic regimen effectively channels more methane-derived carbon into extracellular acetate. Our findings underscore that redox conditions critically shape methanotrophic metabolism, influencing both the biological methane cycle and the global climate.

|
Scooped by
mhryu@live.com
February 4, 1:57 PM
|
The research on synthetic methylotrophic bacteria for one-carbon (C1) feedstock assimilation has garnered substantial interest and is regarded as the forefront of biomanufacturing advancements. Nevertheless, the effective utilization of C1 feedstocks faces challenges due to inadequate tolerance toward C1 compounds. This study elucidates that the buildup of formaldehyde causes severe DNA–protein cross-linking (DPC), and thus hampers growth and methanol assimilation in E. coli. To tackle this issue, we exploited a metalloproteinase, SpWss1, from Schizosaccharomyces pombe. By fine-overexpressing SpWss1 in the E. coli genome, we were able to alleviate DPC damage and enhance formaldehyde tolerance. Remarkably, the engineered strain displayed a 10-fold increase in the amount of methanol assimilated (142 mM) compared to that of the control strain lacking SpWss1 (14 mM). Moreover, through iterative substrate feeding of methanol and xylose in shake-flask experiments, the genetically modified strain exhibited improved consumption levels, reaching up to 309 mM (∼10 g/L), making it one of the highest methanol-consuming strains among all E. coli strains without adaptive evolution. Additionally, the modified strain significantly enhanced the sustainable production of valuable products, such as triacetic acid lactone and fatty acids, from methanol. Overall, our findings underscore the significant scientific and biotechnological importance of addressing DPC to optimize C1 assimilation, providing valuable insights for sustainable chemistry, engineering, and industrial biotechnology applications.

|
Scooped by
mhryu@live.com
February 4, 1:45 PM
|
Mushroom-forming basidiomycetes are increasingly recognized for their significant potential to remediate polluted environments and mitigate climate change. This review synthesizes evidence positioning mushroom-forming basidiomycetes at the nexus of ecological resilience and a sustainable bioeconomy, highlighting their dual roles in environmental repair and green innovation. Ectomycorrhizal (ECM species) enhance carbon acquisition by plants and long-term soil carbon sequestration; ECM-dominant forests stockpile upto 70% more below-ground carbon than their non-mycorrhizal counterparts. Saprotrophic fungi drive lignocellulose degradation, nutrient cycling, and the stabilization of soil organic matter. Basidiomycetes also play a crucial role in mycoremediation by degrading recalcitrant contaminants (pesticides, hydrocarbons) and immobilizing heavy metals. Furthermore, mycelium-based biomaterials are being developed as green-technology alternatives to plastics and synthetic foams, reflecting the growing commercialization of fungal biotechnology, as evidenced by the global mycelium material industry projected to exceed USD 5 billion by 2032. The intersection of ecological function and economic value positions mushrooms at the forefront of the circular bioeconomy. However, challenges remain, including production scalability, environmental sensitivity, and economic viability. Addressing these challenges through interdisciplinary research could unlock the full potential of fungi as nature-based climate solutions.

|
Scooped by
mhryu@live.com
February 4, 1:24 PM
|
Komagataella pastoris is extensively used as a microbial cell factory for the production of recombinant proteins and high-value compounds. However, tightly controlled promoter systems responsive to safe and economical inducers are required for precise metabolic and pathway engineering in this yeast species. Cumate-inducible promoters are an ideal choice due to the safety and low cost of cumate. In this study, we systematically optimised the insertion sites of the CuO operator sequence within the strong promoter PGCW14 to isolate a high-activity variant that we designated as PGCWCuO03. To fine-tune the expression of the repressor protein CymR, we developed a truncated promoter of PGAP, designated as PGAP200. Based on the optimal promoter PGCWCuO03 and the CymR expression unit, we constructed a robust CymR/CuO-mediated cumate-inducible promoter, designated as Pgc, in K. pastoris. Pgc demonstrated outstanding induction properties, resulting in an approximately 11-fold increase in target protein production following induction. Promoter substitution assays validated the effectiveness of Pgc in temporal gene expression control, highlighting the significant potential of this promoter for both basic research and industrial bioprocessing applications in synthetic biology and biotechnology in K. pastoris.

|
Scooped by
mhryu@live.com
February 4, 1:16 PM
|
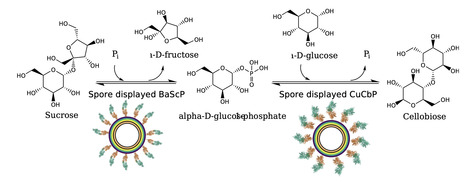
Cellobiose (4-O-β-D-Glucopyranosyl-D-glucopyranose) is an important disaccharide utilized, for example in food and cosmetics. It can be enzymatically synthesized involving two steps from sucrose and glucose, where first the sucrose undergoes phosphorolysis by sucrose phosphorylase, yielding glucose 1-phosphate and fructose. Glucose 1-phosphate is then combined with glucose into cellobiose, releasing phosphate in a reaction catalyzed by cellobiose phosphorylase. To better control and reuse the enzymes in the two main reaction steps, immobilization on Bacillus subtilis spores is a promising approach due to the ease of production and recyclability. Here we describe the display of a sucrose phosphorylase and a cellobiose phosphorylase on B. subtilis spores through fusion with the crust protein CotY, to our knowledge, marking the first use of multiple enzymes directly displayed on the spore surface during sporulation in a reaction cascade. While immobilization had no effect on thermostability, we demonstrate the recyclability of the individual spore variants over four reaction cycles at 45 °C with sucrose phosphorylase maintaining 35% of its initial activity and cellobiose phosphorylase maintaining 65%. Both spore variants were used together to catalyse a reaction cascade in a separated two-pot, as well as in a one-pot reaction. The one-pot reaction achieved a 90% yield with respect to the initially available 40 mM of glucose. The one-pot cascade maintained activity after being recycled five times over the course of 120 hours. Furthermore, we report on improving the reaction yield in the two-pot reaction from 60% to 80% by using calcium to precipitate excess phosphate. In this study we demonstrate that spores are a suitable immobilization platform for multistage reaction cascades. The spores displaying biocatalysts can be recovered and reused over multiple reaction cycles. The immobilization of glycosylic enzymes on spores enables cost-effective, scalable enzyme production on a temperature-resistant carrier that facilitates purification. The potential modularity of this approach adds to the adaptability of the system to different requirements in terms of substrate and product.

|
Scooped by
mhryu@live.com
February 4, 12:57 PM
|
Crops depend on microbial partners for their growth, development, and overall resilience. A pivotal understanding has emerged showing the direct involvement of the root microbiota in regulating the tiller number of rice, a crucial architecture that influences yield. Novel frontiers in microbiological applications for agriculture highlight the profound role of the root microbiota in shaping crop architecture to boost productivity. We propose that improvements in crop production are moving from a genetic perspective on “architecture” to embracing “holobiont architecture.” As such, microbial orchestration provides a dynamic fine-tune function for breeding “architecture-smart crops” characterised by phenotypic plasticity under environmental uncertainty.

|
Scooped by
mhryu@live.com
February 4, 12:44 PM
|
Bacteria use diverse mechanisms to protect themselves against phages. Many antiphage systems form large oligomeric complexes, but how oligomerization is regulated during phage infection remains mostly unknown. Here we demonstrate that the bacterial immunity protein ring-activated zinc-finger RNase (RAZR) assembles into an active, 24-meric ring around the circumference of large ring structures formed by two unrelated phage proteins: a putative recombinase and a portal protein. Each multi-layered, megadalton-scale complex enables RAZR to cleave RNA nonspecifically to inhibit translation and restrict phage propagation. The recognition of unrelated phage proteins that form rings with similar diameters indicates that these proteins not only bind to RAZR but also enforce a geometry crucial to activation. The lack of large ring structures in the host probably prevents auto-immunity and RAZR activation before infection. The infection-triggered oligomerization of RAZR mirrors pathogen-induced oligomerization in eukaryotic innate immune complexes, underscoring a common principle of immunity across biology. An antiphage defence system has an activation mechanism that relies on the sensing of phage-encoded proteins that enforce geometry crucial to activation and are not typically present in non-infected cells.

|
Scooped by
mhryu@live.com
February 4, 12:28 PM
|
Synthetic gene circuits are powerful tools for precisely programming gene expression and introducing novel cellular functions. However, their development and application in plants has lagged behind other systems, due mainly to the limited availability of modular genetic parts. We recently developed a CRISPR interference (CRISPRi)-based synthetic gene circuit system for programming gene expression in plants. Using a robust and high-throughput protoplast-based dual luciferase assay, we demonstrated the development, testing and functionality of these circuits in various plant species. Here we detail the key design principles and considerations for building and testing programmable and reversible CRISPRi-based gene circuits in plants. We also provide detailed procedures for isolating protoplasts from multiple plant species, including Arabidopsis thaliana, Brassica napus, Triticum aestivum and Physcomitrium patens. Furthermore, we provide step-by-step instructions for the 96-well plate-based protoplast transfection assay for testing genetic parts and synthetic circuits, using a dual luciferase assay. The detailed descriptions of these developed systems will enhance the efficiency and reproducibility of the construction, testing, and implementation of synthetic gene circuits in a variety of plant species. This protocol enables the design and testing of CRISPRi-based gene circuits in plants within ~4 weeks. This protocol details the design principles and procedures for building programmable CRISPRi-based gene circuits, and for testing individual components and the assembled circuits in various plant species using a high-throughput protoplast-based assay.
|

|
Scooped by
mhryu@live.com
Today, 12:02 AM
|
Biofilms have emerged as promising biocatalysts due to their distinct structural and functional advantages. Since biofilm dynamics shape biofilm architecture and catalytic performance, engineering strategies to control these dynamics are key to improving biofilm-based catalysis. In this review, we outline the fundamental features and catalytic benefits of biofilms, with a focus on biofilm dynamics. We highlight recent advances in regulatory strategies, from the manipulation of biofilm-associated genes to the design of synthetic circuits based on signaling networks that govern biofilm development. We further discuss current challenges, including limited regulatory efficiency, restricted applicability beyond model organisms, and the need for biofilm functional enhancement. Collectively, these insights position the control of biofilm dynamics as a frontier for advancing next-generation biofilm-based biocatalysis.

|
Scooped by
mhryu@live.com
February 4, 11:35 PM
|
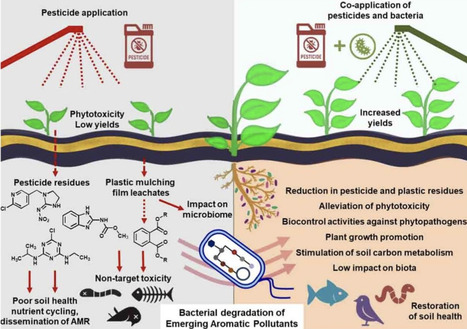
Emerging aromatic pollutants originating from pesticides, including insecticides, herbicides, pesticide additives, surfactants, and mulching films, are increasingly contaminating agro-ecosystems. Their widespread use, recalcitrance, and toxicity pose serious risks to the environment as well as human health. Bacteria possess remarkable potential to degrade these persistent, bioaccumulative, and toxic aromatic pollutants. This review focuses on metabolic fate and eco-physiological traits that bacteria employ for survival and effective degradation of xenobiotics that commonly persist in agricultural fields. It also highlights opportunities and challenges of a holistic approach using bacteria, that is, integrating pollutant biodegradation, plant growth promotion, and biocontrol activities to restore soil health and crop productivity.

|
Scooped by
mhryu@live.com
February 4, 10:44 PM
|
Microbial natural products represent a chemically diverse repertoire of small molecules with major pharmaceutical potential. Despite the increasing availability of microbial genome sequences, large-scale natural product discovery remains challenging because the existing genome mining approaches lack integrated workflows for rapid dereplication of known compounds and prioritization of novel candidates, forcing researchers to rely on multiple tools that requires extensive manual curation and expert intervention at each step. To address these limitations, we introduce LoReMINE (Long Read-based Microbial genome mining pipeline), a fully automated end-to-end pipeline that generates high-quality assemblies, performs taxonomic classification, predicts biosynthetic gene clusters (BGCs) responsible for biosynthesis of natural products, and clusters them into gene cluster families (GCFs) directly from long-read sequencing data. By integrating state-of-the-art tools into a seamless pipeline, LoReMINE enables scalable, reproducible, and comprehensive genome mining across diverse microbial taxa. The pipeline is openly available at https://github.com/kalininalab/LoReMINE and can be installed via Conda (https://anaconda.org/kalininalab/loremine), facilitating broad adoption by the natural product research community.

|
Scooped by
mhryu@live.com
February 4, 2:48 PM
|
Canonical nucleic acids (DNA and RNA) naturally store genetic information with high density and programmability, making them promising candidates for molecular data storage. However, their susceptibility to degradation under harsh conditions, such as extreme pH, nuclease activity, and chemical attack, limits practical applications. In contrast, non-canonical nucleic acids (ncNAs) with natural or synthetic structural modifications exhibit enhanced stability and unique functional potential. This review systematically summarizes the fundamental properties of ncNAs, evaluates their suitability for molecular data storage, and discusses how their distinctive advantages may overcome the intrinsic limitations of canonical nucleic acids while addressing challenges in next-generation storage systems. Canonical nucleic acids, with high storage density, are promising media for data storage but are limited by instability. Here, authors summarise the unique properties and advantages of non-canonical nucleic acids in data storage and highlight key challenges for their practical application.

|
Scooped by
mhryu@live.com
February 4, 2:33 PM
|
Nodule-forming bacteria play crucial roles in plant health and nutrition by providing fixed nitrogen to leguminous plants. Despite the importance of this relationship, how nodule-forming bacteria are affected by plant exudates and soil minerals is not fully characterized. Here, the effects of plant-derived methanol and lanthanide metals on the growth of nitrogen-fixing Rhizobiales are examined. Prior work has demonstrated that select Bradyrhizobium strains can assimilate methanol only in the presence of lanthanide metals; however, the pathway enabling assimilation remains unknown. In this study, we characterize Bradyrhizobium diazoefficiens USDA 110, Bradyrhizobium sp. USDA 3456, and Sinorhizobium meliloti 2011 to determine the pathways involved in methanol metabolism in previously characterized strains, other clades of Bradyrhizobium, and the more distantly related Sinorhizobium. Based on genomic analyses, we hypothesized that methanol assimilation in these organisms occurs via the lanthanide-dependent methanol dehydrogenase XoxF, followed by oxidation of formaldehyde via the glutathione-linked oxidation pathway, subsequent oxidation of formate via formate dehydrogenases, and finally assimilation of CO2 via the Calvin-Benson-Bassham (CBB) cycle. Transcriptomics revealed upregulation of the aforementioned pathways in USDA 3456 during growth with methanol. Enzymatic assays demonstrated increased activity of the glutathione-linked oxidation pathway and formate dehydrogenases in all strains during growth with methanol compared to succinate. 13C-labeling studies confirmed the presence of CBB intermediates and label incorporation during growth with methanol. Our findings provide multiple lines of evidence supporting the proposed XoxF-CBB pathway and, combined with genomic analyses, suggest that this metabolism is widespread among Bradyrhizobium and Sinorhizobium species.

|
Scooped by
mhryu@live.com
February 4, 1:59 PM
|
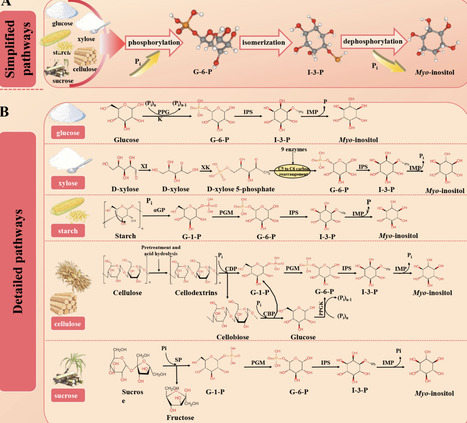
Myo-inositol, a high-value cyclic polyol, is increasingly sought by pharmaceutical, food, feed, and cosmetic industries. This review systematically surveys the latest biotechnological advances poised to replace traditional, high-pollution methods. First, multienzyme cascades that convert renewable carbohydrates─starch, glucose, xylose, cellulose, sucrose─are reviewed, highlighting immobilized reactors, porous microspheres, biomimetic mineralized capsules, and biofilm systems that boost stability, reusability, and space-time yields. Second, microbial cell-factory strategies are examined, covering chassis benchmarking (E. coli, Pichia pastoris, Kluyveromyces marxianus, cyanobacteria), carbon-flux redirection, glucose-glycerol synergistic feeding, and dynamic regulatory circuits. A unified analysis pinpoints recurrent bottlenecks─cofactor imbalance, enzyme thermostability gaps, narrow substrate spectra, product inhibition, and downstream complexity─and distills the targeted fixes discussed in the field, from cofactor regeneration circuits to modular process design. By integrating cutting-edge research with industrial techno-economic indicators, this review offers a comprehensive roadmap for sustainable, cost-competitive myo-inositol biomanufacturing and guides future research toward greener production systems.

|
Scooped by
mhryu@live.com
February 4, 1:50 PM
|
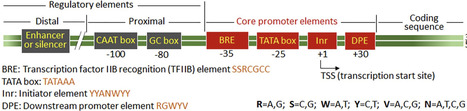
Plant-based expression systems, known as molecular farming, have emerged as sustainable and scalable platforms for producing recombinant proteins used in pharmaceuticals, industrial enzymes, and agricultural products. Among the key determinants of transgene performance, promoter elements play a central role in defining transcriptional strength, specificity, and regulation. This review highlights current advances in promoter engineering tailored for plant systems, encompassing natural, synthetic, hybrid, inducible, and tissue-specific promoters used in stable transgenic plants, transient expression systems, and plant cell cultures. The structural and functional features of promoter elements are discussed, along with strategies to mitigate challenges such as transcriptional silencing, genomic context dependency, and variability cross species and production platform. Emerging synthetic biology tools, such as CRISPR-based transcriptional control, high-throughput screening, and machine learning–assisted promoter design, are enabling the creation of tunable, orthogonal promoters suited for complex multigene expression. As promoter engineering continues to evolve, it remains foundational to advancing plant molecular farming and expanding the role of plants as versatile biofactories for high-value recombinant proteins.

|
Scooped by
mhryu@live.com
February 4, 1:30 PM
|
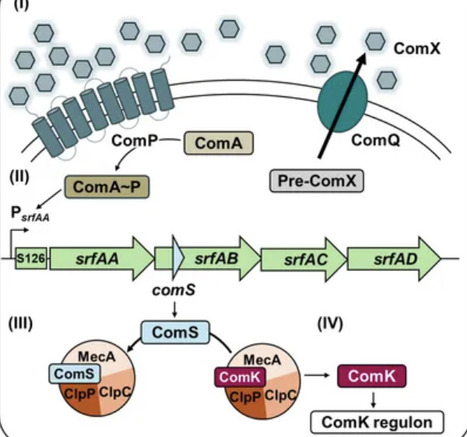
In its natural soil habitat, B. subtilis regularly encounters fluctuating conditions that require adaptive survival strategies, including the production and secretion of antimicrobial compounds. One such compound, surfactin, play a central role in multicellular differentiation processes such as biofilm formation, swarming, and competence development. Competence and surfactin biosynthesis are transcriptionally co-regulated via the quorum sensing-mediated activation of the srfAABCD operon, which contains comS in a distinct ORF overlapping with srfAB. This study aimed at uncoupling competence from surfactin production by introducing targeted stop mutations in comS to selectively disrupt competence without affecting surfactin synthesis. For this, we introduced single nucleotide polymorphisms (SNPs) that preserved the srfAB codons, while simultaneously introducing a premature stop codon in comS. The effects on competence development were assessed using luciferase-based reporter assays monitoring the ComS-dependent expression of comK and comGA expression. Surfactin production was analyzed by mass spectrometry imaging and phenotypic assays examining the impact on multicellular behavior. Our findings demonstrate that the generated point mutations severely reduce competence gene expression, measured via PcomKand PcomGAactivity, to levels comparable with a full comS deletion, while leaving multicellular behaviors such as biofilm and pellicle formation, as well as swarming and sliding motility, unaffected. Thus, ComS is specifically essential for competence development but dispensable for other surfactin-mediated multicellular processes and not involved in structuring biofilms. Taken together, our results demonstrate that it is possible to genetically decouple competence from other developmental pathways in B. subtilis.

|
Scooped by
mhryu@live.com
February 4, 1:21 PM
|
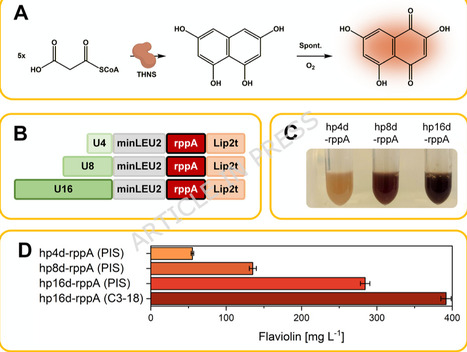
Yarrowia lipolytica is an emerging host for producing acetyl-CoA– and malonyl-CoA–derived chemicals. However, most processes rely on yeast nitrogen base (YNB), a historical formulation with poorly controlled trace metal content. This variability impairs metabolic performance, limits reproducibility, and complicates process transfer. Commercial YNB batches differed markedly, causing 1.5–2-fold variation in growth and docosahexaenoic acid (DHA) production. We developed a malonyl-CoA–responsive flaviolin reporter strain and combined it with a structured Design of Experiments (DoE) workflow to systematically re-engineer YNB mineral composition. Dissection of all 20 YNB components revealed that vitamins are dispensable under the tested conditions, whereas a small subset of salts and trace elements - particularly ZnSO4, FeCl3, KH2PO4, MgSO4, CaCl2, and CuSO4 - dominantly shape precursor availability and product formation. One-factor-at-a-time (OFAT), factorial, steepest ascent, and central composite designs converged in an optimized synthetic mineral medium assembled entirely from individual salts and trace metals. This formulation increased flaviolin titers to 1.41 ± 0.08 g L-1, a more-than threefold improvement over commercial YNB, while ensuring high reproducibility. Key mineral interventions also translated to complex pathways: omission of ZnSO4 increased PUFA titers by 7.6-fold (docosapentaenoic acid, DPA) and 58-fold (eicosapentaenoic acid, EPA) and enhanced DHA formation in independent production strains. The defined formulation substantially reduces cost and eliminates batch-to-batch variability inherent to commercial YNB powders. Our results establish mineral balancing as a major yet underused lever for improving acetyl-CoA– and malonyl-CoA–derived production in Y. lipolytica and demonstrate a generalizable, model-guided workflow for creating simplified, reproducible, and cost-efficient synthetic media for non-conventional yeast cell factories.

|
Scooped by
mhryu@live.com
February 4, 1:00 PM
|
Numerous Gram-negative bacteria possess N-acyl-L-homoserine lactone (AHL)-mediated quorum-sensing (QS) systems that regulate the activation of specific genes. Burkholderia plantarii causes rice seedling blight by producing the phytotoxin tropolone. In this study, we investigated multiple AHL-type QS systems in B. plantarii MAFF 301723T and their involvement in virulence regulation. MAFF 301723 harbors three AHL-mediated QS systems, designated plaI1/plaR1, plaI2/plaR2, and plaI3/plaR3. The plaI1/plaR1 system, which produces N-octanoyl-l-homoserine lactone, is functional and essential for swarming motility. When forced expression of plaI2 induces the biosynthesis of 3-OH-C10-HSL, it was suggested that expression is rarely observed in wild-type MAFF 301723. The plaI3 gene directs the synthesis of the putative C16:2-HSL, which is a rare AHL bearing two double bonds in the hexadecanoyl chain that has not been previously reported in Burkholderia spp. The plaI3/plaR3-QS system is crucial for tropolone production. These findings suggest that multiple QS systems collectively contribute to the complex virulence regulation of B. plantarii, thereby providing new insights into the development of QS-targeted biocontrol strategies for agriculture.

|
Scooped by
mhryu@live.com
February 4, 12:55 PM
|
Photoreceptor proteins regulate fundamental biological processes such as vision, photosynthesis and circadian rhythms. A large photoreceptor subfamily uses vitamin B12 derivatives for light sensing, contrasting with the well-established mode of action of these organometallic derivatives in thermally activated enzymatic reactions. The exact molecular mechanism of B12 photoreception and how this differs from the thermal pathways remains unknown. Here we provide a detailed description of photoactivation in the prototypical B12 photoreceptor CarH from nanoseconds to seconds, combining time-resolved and temperature-resolved structural and spectroscopic methods with quantum chemical calculations. Building on the crystal structures of the initial tetrameric dark and final monomeric light-activated states, our structural snapshots of key intermediates in the truncated B12-binding domain illustrate how photocleavage of a cobalt–carbon (Co–C) bond within the B12 chromophore adenosylcobalamin triggers a series of structural changes that propagate throughout CarH. Breakage of the photolabile Co–C5′ bond leads to the formation of a previously unknown adduct that links the C4′ position of the adenosyl moiety to the Co ion and can subsequently be cleaved thermally over longer timescales to allow release of the adenosyl group, ultimately causing tetramer dissociation. This adduct, which differentiates CarH from thermally activated B12 enzymes, steers the photoactivation pathway and acts as the molecular bridge between photochemical and photobiological timescales. The biological relevance of our study is corroborated by kinetic data on full-length CarH in the presence of DNA. Our results offer a spatiotemporal understanding of CarH photoactivation and pave the way for designing B12-dependent photoreceptors for optogenetic applications. Spatiotemporal insight into photoactivation of the prototypical B12 photoreceptor CarH is revealed across nine orders of magnitude in time, identifying a transient adduct that distinguishes it from thermally activated B12 enzymes.

|
Scooped by
mhryu@live.com
February 4, 12:32 PM
|
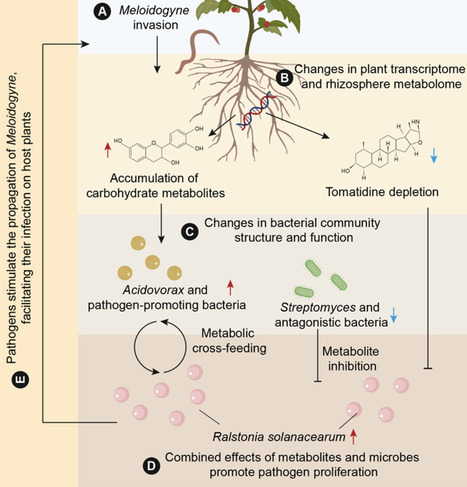
Root-knot nematodes cause substantial crop losses by compromising plant immunity and facilitating invasion by soil-borne bacterial pathogens, yet the mechanisms underlying nematode-facilitated co-infection remain poorly understood. Here, we quantify the global prevalence of nematode-pathogen co-infection and integrate multi-omic analyses across greenhouse and in vitro experiments. We show that nematode invasion activates plant defense gene expression but concurrently disrupts rhizosphere homeostasis, resulting in microbiome dysbiosis that overrides host resistance. Meloidogyne invasion induces pronounced metabolic reprogramming, characterized by depletion of tomatidine and accumulation of carbohydrate metabolites such as galactose. These shifts selectively suppress Streptomyces-dominated antagonistic microbiota while enriching Acidovorax, which exhibits nutritional synergy with Ralstonia. Using synthetic microbial community transplantation, we demonstrate a functional transition from pathogen-suppressive to pathogen-permissive bacteriomes following nematode invasion. Together, our findings reveal how nematodes and bacterial pathogens cooperatively subvert plant-microbe metabolic signaling to undermine rhizosphere immunity, highlighting actionable targets for microbiome-based disease control.
|
 Your new post is loading...
Your new post is loading...
























laub mt, Despite harboring a HEPN domain and functioning through an abortive infection mechanism, RAZR was not toxic to E. coli when overproduced. Thus, we hypothesized that RAZR must be activated by some phage factor (or factors) during infection. To identify these triggers, we isolated escape mutants of SECΦ27 that overcome RAZR defence. All escape phage clones contained point mutations leading to a single amino acid substitution (S19R, S31R or D41Y) in Gp77.