 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:41 AM
|
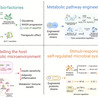
Metabolic disorders, including type 2 diabetes mellitus, obesity, metabolic dysfunction-associated steatohepatitis, etc., are an escalating global health challenge with substantial clinical and socio-economic burdens. Conventional pharmacological therapies are constrained by short-term efficacy, instability, and energy-intensive production. This review aims to evaluate how synthetic biology enables engineered probiotics and microbial systems as self-regulating, living therapeutics that integrate precision treatment with sustainable, bio-based production. Such systems can secrete therapeutic molecules, degrade harmful metabolites, remodel host metabolic microenvironments, and respond to physiological signals to achieve adaptive, feedback-controlled interventions. By exploiting versatile microbial hosts and low-energy fermentation, they minimize chemical waste and carbon footprint. We also discussed preclinical studies demonstrating restored glucose and lipid homeostasis, modulation of appetite, and attenuation of inflammation. Collectively, our review highlights that synthetic biology exemplifies a transformative, sustainable paradigm for metabolic disease management.

|
Scooped by
mhryu@live.com
Today, 12:34 AM
|
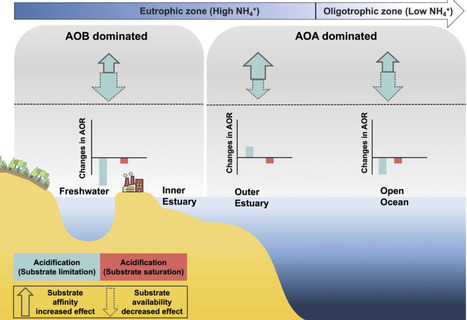
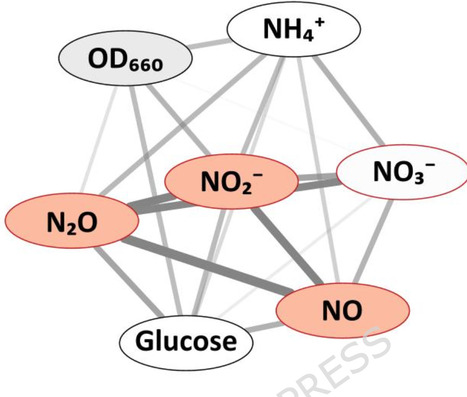
Ammonia oxidation, a critical nitrogen cycle process, exhibits contradictory responses to aquatic acidification, and the underlying mechanism remains unresolved. Here, through pH manipulation experiments across diverse ecosystems and with a model ammonia-oxidizing archaea species, Nitrosopumilus maritimus strain SCM1, we discover a unifying adaptive mechanism: acidification triggers a compensatory increase in substrate affinity in ammonia-oxidizing microorganisms. This enhancement counteracts the reduction in ammonia availability, with the magnitude of increase being significantly greater in ammonia-oxidizing archaea than in ammonia-oxidizing bacteria. Consequently, in ammonia-oxidizing archaea-dominated systems, this adaptation can sustain or even stimulate oxidation rates under moderate acidification, while ammonia-oxidizing bacteria-dominated systems experience a decline. By incorporating this affinity response into models, we accurately reconcile prior disparate field observations. We thus establish the regulation of substrate affinity as a key determinant of microbial resilience, providing a framework for predicting nitrogen cycle dynamics under future acidification. This study shows that acidification increases substrate affinity in ammonia-oxidizing microbes, partly offsetting reduced ammonia availability. Stronger responses in archaea than bacteria explain contrasting ecosystem reactions.

|
Scooped by
mhryu@live.com
Today, 12:04 AM
|
Calcium oxalate stones comprise greater than 70% of all kidney stones. In the current conceptual framework, the initial stone nidus is thought to include the aggregation of inorganic crystallites, the formation of which is favored by elevated concentrations of dissolved constituents. Here, we show that this highly prevalent stone type comprises a form of organic–inorganic polycrystalline biocomposite with integrated bacterial biofilms. Evidence from electron microscopy and fluorescence microscopy reveal the unanticipated internal structure of kidney stones from human patients, where bacterial biofilms are intercalated between polycrystalline mineral layers, even in stones identified as “noninfectious” clinically, including those in patients without underlying urinary tract infections. We observe similar bacterial biofilm architectures on the surfaces of stone fragments obtained due to lithotripsy, suggesting that bacteria are intrinsic to the process of nephrolithiasis. Crystallites proximal to biofilm layers exhibit significantly smaller grain sizes, which indicate a larger local concentration of nucleation sites. Staining reveals that biofilm areas of these stones are enriched with bacterial DNA. That bacteria are now observed so broadly in kidney stones (including even in less prevalent struvite stones) may be conceptually salient: Based on the evidence adduced here, we propose a model in which the urine-rich environment of the kidney can impinge on bacterial calcium homeostasis and amplify bacterial production of nucleation templates such as extracellular DNA. The resultant counterion condensation intrinsic to polyelectrolytes charged beyond the Manning criterion (such as DNA) drastically enhances the probability of heterogeneous nucleation, thereby amplifying calcium oxalate stone formation.

|
Scooped by
mhryu@live.com
January 27, 11:47 PM
|
Methanogenesis is a crucial component of Earth's carbon cycle and a source of methane for biofuel production. The presence of higher energy electron acceptors, such as iron(III) oxides and quinones, is believed to significantly impact methanogenesis. This study investigated the physiological and proteomic responses of the type I Methanosarcina, M. barkeri, to the artificial quinone 9,10-anthraquinone-2,7-disulfonate disodium (2,7-AQDS), using H2/CO2 as substrates. Our findings revealed that during 2,7-AQDS reduction, cellular growth ceased. The lack of energy conservation was associated with direct inhibition of both methanogenesis and CO2 utilization, corroborated by a significant downregulation of the enzymes involved in this metabolic pathway. Furthermore, the significant upregulation of specific subunits of the reversible Ech hydrogenase suggests that this enzyme redirects electrons from H2 towards the most energetically favorable reaction (2,7-AQDS reduction), rather than the reduction of ferredoxin, which is a highly energy-demanding process, essential for initiating the CO2 reduction pathway. Additionally, it is conceivable that Ech homologues in other hydrogenotrophic methanogens also participate in the reduction of higher energy-yielding electron acceptors. These findings provide novel insights into how quinones, particularly in their oxidized state, directly impact methanogenesis, thereby influencing both artificial and natural methanogenic environments.

|
Scooped by
mhryu@live.com
January 27, 11:17 PM
|
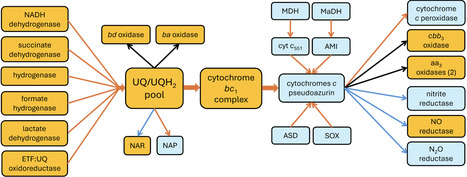
Paracoccus denitrificans is a metabolically versatile alphaproteobacterium first isolated in 1910 by Martinus Beijerinck. Similarities between the aerobic respiratory chain and membrane composition of P. denitrificans and those of eukaryotic mitochondria have stimulated the use of P. denitrificans as a model for oxidative phosphorylation. The organism has also been used extensively as a model for studies of denitrification, cytochrome c biogenesis, lithotrophy using thiosulphate as a source of energy, methylotrophy, and carbon metabolism more broadly. Through the application of structural biology and modern genome-based approaches, work on P. denitrificans continues to make significant contributions across multiple areas of microbiology.

|
Scooped by
mhryu@live.com
January 27, 11:05 PM
|
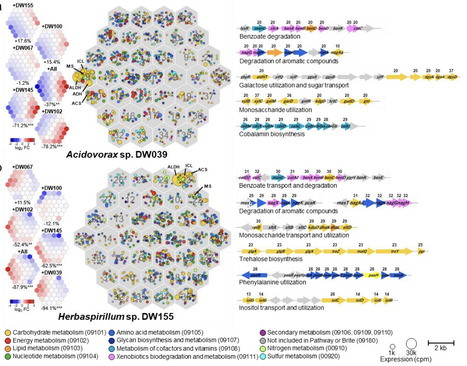
Plants are colonized by characteristic microbiomes that help maintain plant health and function. However, the mechanisms that make these communities assemble in a consistent, repeatable manner remain poorly understood. Here, we show how microbial metabolic strategies for host-derived substrates drive plant microbiome assembly, using a six-member synthetic community syncom that captures the taxonomic diversity of natural duckweed microbiome. Contrary to expectations of metabolic niche partitioning, five of six strains adopt similar metabolic states when colonizing the host alone, consistent with the preferential use of a shared substrate, acetaldehyde. In contrast, during competition with specific community members, these strains consistently switch toward alternative substrates, including sugars and aromatics. Strikingly, this metabolic switching accompanies all negative interspecies interactions observed in the community, indicating that a single class of metabolic response dominates the inhibitory interaction network organizing community structure. By comparison, although metabolite cross-feeding is widespread, its contribution to facilitative interactions depends on the background of resource competition. Together, these findings highlight competition-induced metabolic switching as a primary driver of microbial interaction networks, and provide a cross-scale account of how microbial metabolic strategies underpin the reproducible assembly of plant microbiomes.

|
Scooped by
mhryu@live.com
January 27, 7:22 PM
|
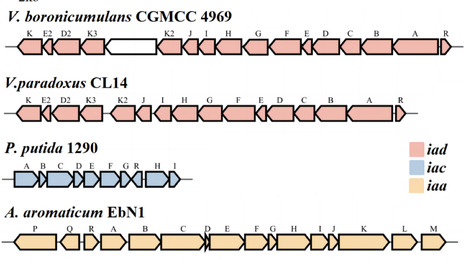
Plant growth-promoting rhizobacteria (PGPR) of the genus Variovorax facilitate plant growth through beneficial microbe-plant interplay. Unlike most PGPR, Variovorax boronicumulans CGMCC 4969 utilizes indole-3-acetonitrile as a precursor to generate indole-3-acetic acid (IAA), which is subsequently metabolized by itself. In this study, IAA enhanced the growth of V. boronicumulans CGMCC 4969 in minimal salt medium (MSM), whereas it inhibited bacterial growth when glucose was added to the MSM broth. IAA was rapidly degraded within 12 h in MSM broth despite glucose appeared or not. Notably, in LB broth, the cell growth was significantly inhibited by IAA concentration beyond 1 mmol/L, while the IAA degradation capability of CGMCC 4969 was significantly increased following exposure to IAA-dosed LB medium. V. boronicumulans CGMCC 4969 degraded IAA to yield a new intermediate 3-hydroxy-anthranilate. An iad gene cluster was identified in V. boronicumulans CGMCC 4969, and co-expression of the iadD and iadE genes endows E. coli with the capacity to degrade IAA. This degradation efficiency is augmented when the iadC gene is expressed simultaneously. Subsequent proteomics and bioinformatics analyses highlighted that the addition of IAA induced a significant up-regulation of ABC transporter proteins, in particular IadK3 and IadK2. Interestingly, there was also a significant increase in protein expression associated with group-sensing metabolism. Collectively, this research helps our understanding of the intricate regulatory mechanisms of IAA within Variovorax own metabolism and expands our knowledge of its complex role in plant-microbe interactions. Key points The iad gene cluster degraded IAA to a previously uncharacterized intermediate.Adding IAA during the cell culture period enhances IAA-degrading activity.Proteomics defined the adaptive response to IAA.

|
Scooped by
mhryu@live.com
January 27, 2:18 PM
|
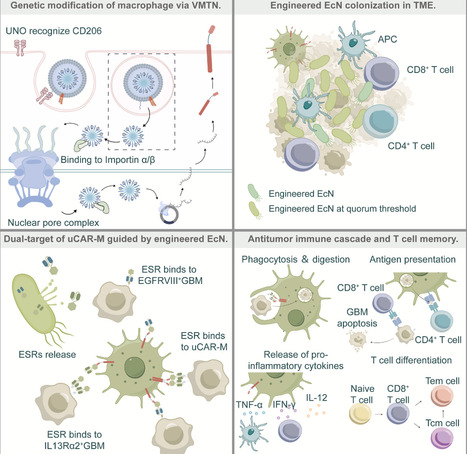
Glioblastoma (GBM) remains a highly lethal form of cancer due to its molecular heterogeneity and the immunosuppressive microenvironment surrounding the tumor. Here, we report a modular immunotherapy platform characterized by its flexibility to simultaneously target multiple antigens. Specifically, we utilize engineered E. coli Nissle to colonize tumors and produce bispecific engagers that simultaneously target EGFRvIII and interleukin (IL)-13Rα2. These tags direct in situ-reprogrammed chimeric antigen receptor (CAR) macrophages, which are edited using nanoparticles and delivered within a shear-thinning hydrogel, to execute targeted phagocytosis. This probiotic-macrophage crosstalk eliminates tumor cells while converting protumor M2 macrophages into immunostimulatory M1 effectors. In aggressive orthotopic GBM mouse models, this strategy achieves 83% survival at the 120-day endpoint, representing a 5-fold improvement over single-target controls and establishing durable immunological memory that effectively combats recurrence. By functioning as multifunctional immune hubs, this platform offers a versatile framework designed to overcome the antigenic complexity of solid tumors.

|
Scooped by
mhryu@live.com
January 27, 1:59 PM
|
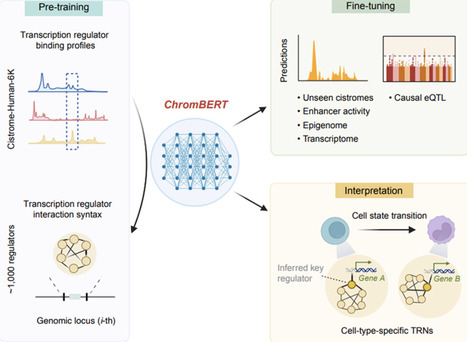
Gene expression is shaped by transcriptional regulatory networks (TRNs), where transcription regulators interact within regulatory elements in a context-specific manner. Deciphering context-specific TRNs has long been constrained by the severe sparsity of cell-type-specific chromatin immunoprecipitation sequencing (ChIP-seq) profiles. Here, we present ChromBERT, a foundation model pre-trained on large-scale human ChIP-seq datasets covering ∼1,000 transcription regulators. ChromBERT learns the genome-wide syntax of regulatory cooperation and generates interpretable TRN representations. After prompt-enhanced fine-tuning, it outperforms existing methods for imputing unseen cistromes. Moreover, lightweight fine-tuning on cell-type-specific downstream tasks adapts the TRN representations to capture regulatory effects and dynamics within any given cellular context. The resulting context-specific representations can then be interpreted to infer regulatory roles of transcription regulators underlying these cell-type-specific regulatory outcomes without requiring additional ChIP-seq experiments. By overcoming the limitations of sparse transcription regulator data, ChromBERT significantly enhances our ability to model and interpret transcriptional regulation across a wide range of biological contexts.

|
Scooped by
mhryu@live.com
January 27, 1:00 PM
|
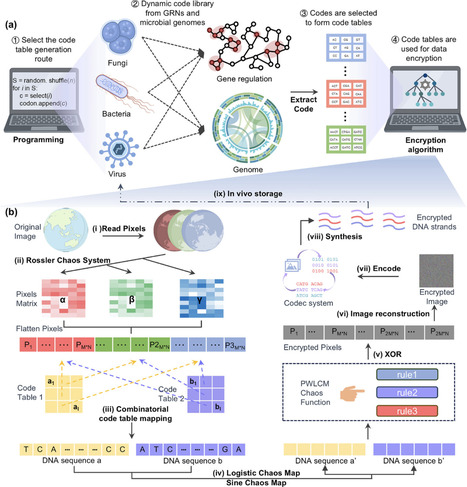
DNA is a promising medium for next-generation data storage because of ultrahigh information density and stability. DNA storage within living organisms presents further advantages, such as self-replication, compactness, and concealment. Early efforts primarily developed predetermined methods for encoding and decoding data using in vivo DNA sequences. However, these methods may pose a security risk while opening a clear channel for potential data access and breaches. To address these challenges, we propose a unified paradigm, integrated computational–biological programming (ICBP), by exploiting the intrinsic digital characteristics within computational and microbial systems. ICBP involves the construction of dynamic code tables from gene regulatory networks or complete genomes across diverse species, expanding the key space by more than 100 orders of magnitude compared with existing methods. The encryption algorithm in ICBP benefits from DNA encoding, computing, and computational operations, leading to superior encryption quality and resistance to brute force and statistical attacks. Furthermore, we demonstrated the practical utility of ICBP via the successful encryption, microbial storage, and decryption of digital files within living systems, achieving 100% data recovery after 100 generations of replication. By combining computational logic with the biological complexities of living systems, the ICBP offers a transformative strategy for secure DNA data storage.

|
Scooped by
mhryu@live.com
January 27, 10:45 AM
|
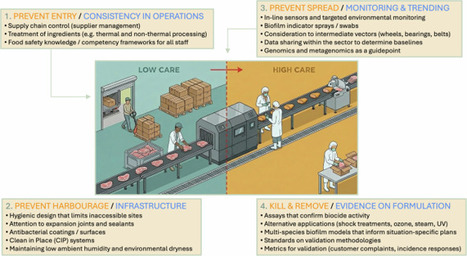
Food safety risks are controlled in agrifood settings by reducing the microbial burden in food ingredients and food production environments. Hygiene programmes involve an incremental implementation of chemical treatments (e.g., disinfectants) and engineering controls (e.g., elimination of susceptible harbourage sites). The strategies to disrupt the presence and transmission of microbial risks to foods are being refined by advanced microbiology and genomics that provide actionable evidence on the precise nature of local ecologies.

|
Scooped by
mhryu@live.com
January 27, 10:36 AM
|
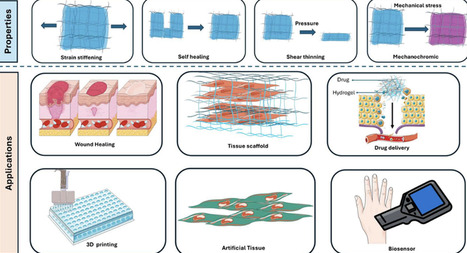
Mechanoresponsive biomaterials are a revolutionary class of materials designed to respond dynamically to mechanical stimuli, providing tissue engineering and regenerative medicine with precise control over biological processes. Through processes including supramolecular interactions, strain stiffening, and force-induced conformational changes, these materials, which include hydrogels, elastomers, and piezoelectric composites, imitate the mechanics exclusive to biological tissues. Additionally, mechanoresponsive systems improve drug delivery by releasing drugs in response to pH changes or mechanical strain using various materials, including magnetic scaffolds and ultrasound-triggered micelles. Despite advancements in numerous arenas of biological sciences, problems with clinical translation, scalability, and long-term biocompatibility still exist. New developments combine technologies like 4D bioprinting to create dynamic, patient-specific scaffolds and artificial intelligence (AI)-assisted design to maximize material qualities. To achieve material innovation with the desired level of biological complexity, future initiatives should focus on multifunctional platforms that combine mechanical, electrical, and biochemical inputs at an advanced level. This review dives into several aspects of mechanoresponsive biomaterials by navigating through the fabrication methods, underlying principles, inception of these in biomedical applications, and progression through the current research settings.

|
Scooped by
mhryu@live.com
January 27, 10:24 AM
|
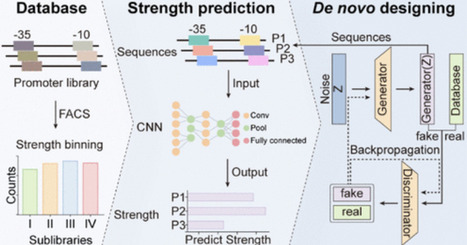
Promoters are essential in transcriptional regulation, with the −10 and −35 boxes playing a critical role in determining their strength. Modulating these regions can effectively fine-tune promoter strength. However, the lack of a clear quantitative relationship between sequence composition and transcriptional output impedes the rational design of promoters. To address this, we developed a synthetic promoter library by varying RNA polymerase binding energies at the −10 and −35 boxes. The library was partitioned into four sublibraries with expression strengths spanning an 80-fold range. Using fluorescence-activated cell sorting FACS followed by sequencing, we identified 20,799 distinct promoters. Analysis of this library uncovered distinct sequence-activity patterns, including a small subset of −35 box sequences that consistently conferred high transcriptional output across diverse −10 partners. Based on this, we developed an artificial intelligence platform that integrates a convolutional neural network for strength prediction (Pearson’s r = 0.84) with a balanced generative adversarial network incorporating a gradient penalty for de novo promoter design. By coupling these models, we achieved a precise design of promoters with user-defined strengths (r = 0.85), establishing a bidirectional framework that links −10/–35 boxes to transcriptional activity through deep learning. This study expands the sequence diversity of functional −10 and −35 boxes in E. coli, provides a predictive platform for rational promoter engineering, and deciphers combinatorial motif interactions governing transcriptional regulation.
|

|
Scooped by
mhryu@live.com
Today, 12:38 AM
|
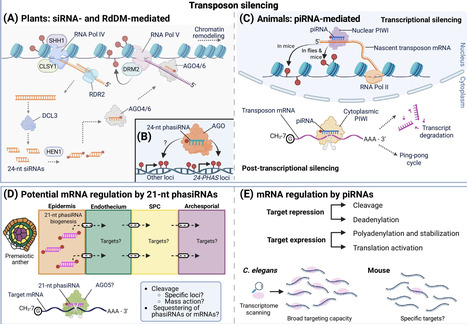
Small RNAs are fundamental to gene expression regulation, with specialized classes playing critical roles in reproduction. This review compares animal PIWI-interacting RNAs (piRNAs) and plant reproductive phased small interfering RNAs (phasiRNAs), which show remarkable similarities. Both originate from Pol II-transcribed precursors but have distinct biogenesis pathways. piRNA processing in metazoans is Dicer-independent, involving PIWI-clade proteins for amplification via ‘ping-pong’ and phased cleavage. Reproductive phasiRNAs are Dicer-dependent and are initiated by miRNA-guided cleavage to generate phased sRNAs. A well-defined piRNA function is transposon silencing, but roles for nontransposon-targeting piRNAs and most reproductive phasiRNAs remain unresolved. Comparing these independently evolved systems reveals common strategies for reproductive success and highlights key unresolved questions regarding their molecular targets, functions, and evolutionary pressures that shaped them.

|
Scooped by
mhryu@live.com
Today, 12:07 AM
|
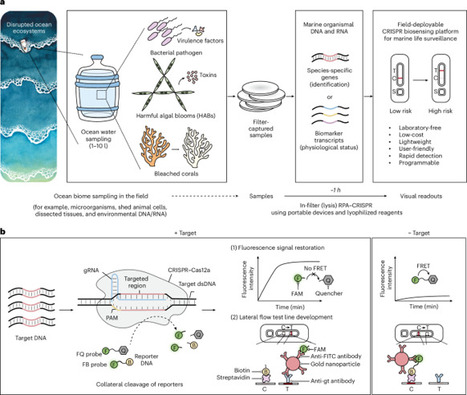
Ocean ecosystems are undergoing accelerating disruption from human impacts such as climate change. Warming ocean temperatures drive pathogenic outbreaks, increase harmful algal blooms and cause coral stress. These can have serious consequences for marine ecosystems, human health and the aquaculture industry, representing a critical One Health issue. Monitoring key marine species offers valuable insights, but current methods are resource-intensive, low-resolution and unsuitable for frequent deployment. Here we introduce a low-cost, field-deployable CRISPR biosensing platform for detecting marine organismal DNA and RNA. Harnessing the programmability of CRISPR diagnostics for environmental biosurveillance, we demonstrate versatility across three climate-linked indicators: Vibrio spp., Pseudo-nitzschia spp. and heat-stressed corals. Portable 3D-printed processor and incubator devices enable direct processing of filter-captured samples with temperature control. Field readiness is reinforced by lyophilized reagents, lateral flow readouts, dropper-based handling and a two-step multiplexed workflow, delivering results within 1 hour without laboratory instruments. Benchmarking with authentic pathogens and environmental seawater confirmed seawater tolerance and robust detection of 108 colony-forming units per filter of Vibrio pathogens, equivalent to 102 copies per microlitre for 1 litre of filtered sample. This decentralized platform reduces barriers to routine monitoring and can provide early warnings of ecosystem disturbances, while supporting One Health initiatives in the marine space. Human impacts on marine ecosystems are increasing the likelihood of pathogenic outbreaks, harmful algal blooms and coral stress. Here the authors develop a CRISPR biomonitoring tool that can help detect key marine species that are important to public health, the aquaculture sector and marine ecosystems.

|
Scooped by
mhryu@live.com
Today, 12:00 AM
|
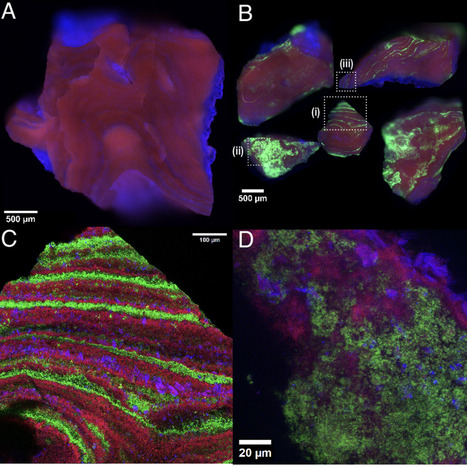
Nucleoid compaction in bacteria is commonly attributed to cytoplasmic crowding, DNA supercoiling, and nucleoid-associated proteins (NAPs). In most bacterial species, including E. coli, these effects condense the chromosome into a subcellular region and largely exclude ribosomes to the surrounding cytoplasm. In contrast, many Mycoplasma—including the Mycoplasma-derived synthetic cell JCVI-Syn3A—exhibit a cell-spanning nucleoid with ribosomes distributed throughout. Because Mycoplasma are evolutionarily distant from model bacteria like E. coli and have undergone extensive genome reduction, Syn3A is a natural testbed for genotype-to-‘physiotype’-to-phenotype, in which genome-encoded composition reshapes cell-scale organization. Here we show that this organization can arise from Syn3A’s unusually high abundance of positively charged proteins. We develop a coarse-grained model that explicitly and physically represents a sequence-accurate chromosome together with ribosomes and cytoplasmic proteins at physiological size, charge, and abundance. With DNA and ribosomes alone, the cell-spanning nucleoid relaxes toward a compacted state that sterically excludes ribosomes, indicating missing physics beyond polymer mechanics and excluded volume. When we include electrostatic interactions by assigning effective charges to each biomolecule, positively charged proteins dynamically enrich around ribosomes and DNA, partially screening ribosome–DNA repulsion. This charge shielding enables ribosomes to penetrate the nucleoid mesh and stabilizes a cell-spanning nucleoid consistent with experiment. This behavior is robust across parameter sweeps: DNA stiffness, heterogeneous mesh size, and crowding favor compaction, whereas electrostatics and size polydispersity promote expansion, with consequences for migration pathways within the nucleoid and thus transcription–translation dynamics. The framework is parameterized directly from genomic and proteomic composition and is transferable to other bacteria.

|
Scooped by
mhryu@live.com
January 27, 11:44 PM
|
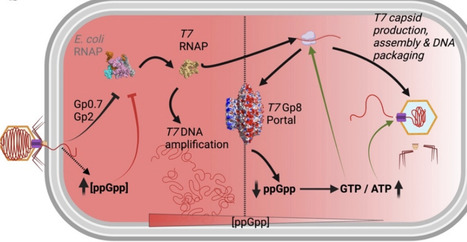
Bacteriophages must overcome host stress responses to replicate efficiently, yet how they engage with these pathways remains poorly understood. The bacterial stringent response (SR), mediated by the alarmone (p)ppGpp, globally suppresses metabolism and limits biosynthetic capacity, posing a potential barrier to phage infection. Here, we show that bacteriophage T7 directly counteracts the SR through its portal protein, Gp8. Using complementary genetic, biochemical, and infection assays, we demonstrate that Gp8 physically interacts with the (p)ppGpp synthetases RelA and SpoT, selectively inhibiting their synthetase activities and reducing intracellular (p)ppGpp levels during late infection. Disruption of this interaction delays host cell lysis and compromises phage replication, while elevated host (p)ppGpp levels impair infection efficiency. Remarkably, portal proteins from diverse E. coli phages share similar structural and functional properties, suggesting a conserved viral strategy to suppress the stringent response. Together, our findings uncover an unexpected regulatory role for phage portal proteins and reveal a conserved mechanism by which phages modulate bacterial stress physiology to optimize replication.

|
Scooped by
mhryu@live.com
January 27, 11:10 PM
|
Functionalizing bacteria with magnetic nanoparticles (MNPs) is a common route toward magnetically responsive bacterial microrobots. However, existing strategies are often limited by low functionalization efficiency, weak binding, poor reproducibility, and nonspecific interactions. Here, we present a robust, specific, and reproducible magnetic functionalization platform based on the self-labeling protein HaloTag, yielding magnetic bacterial microrobots termed HaloBots. Using E. coli as an engineering host, HaloTag was displayed on the outer membrane via the Lpp OmpA anchoring system, with the assistance of a Long and Flexible (LF) peptide linker. Optimization of genetic engineering vectors and colony purification enabled robust and uniform HaloTag display across bacterial populations. In addition, MNPs were modified with chloroalkane PEG ligands to enable site-specific covalent conjugation with HaloTag. Tuning the ligand density on MNPs revealed a critical balance between ligand accessibility and surface charge for achieving efficient and specific attachment. Consequently, the resulting HaloBots exhibited stable MNP conjugation and reliable magnetic actuation. Collectively, this work establishes a modular and tunable strategy for engineering magnetically responsive bacterial microrobots.

|
Scooped by
mhryu@live.com
January 27, 7:28 PM
|
Random variation in reproductive success--genetic drift--profoundly shapes genetic diversity and evolutionary trajectories. The strength of drift depends on the variance in descendant number, σd2, which governs key evolutionary outcomes: for instance, the establishment probability of a beneficial mutation scales inversely with σd2. However, whether σd2 itself evolves over long timescales has remained unclear, because allele-frequency fluctuations depend on drift only through the effective population size, Ne = N/σd2, which blends census population size with descendant-number variance. Here, we disentangle these components by using model-based Bayesian inference combined with joint tracking of (i) frequency fluctuations of neutrally barcoded lineages and (ii) census population sizes across growth cycles in the E. coli Long-Term Evolution Experiment. Analyzing 33 clones spanning the ancestor through 50,000 generations in two replicate populations (Ara-2 and Ara+2), we find that the strength of genetic drift evolved markedly--and divergently--between the two replicate populations. Both census size and σd2 changed substantially through time, with most variation in Ne driven by shifts in σd2 rather than census size. After approximately 2,000 generations, the σd2 of the two populations diverged sharply: Ara+2 generally remained close to a bottleneck-only null expectation, whereas Ara-2 exhibited 1.5-5x stronger drift, consistent with an evolved increase in stochasticity during growth. Because establishment probability scales as s/σd2, a beneficial mutation of given effect is roughly twice as likely to establish in Ara+2 as in Ara-2. Our results demonstrate that the key parameter governing genetic drift can itself evolve, with direct consequences for adaptation.

|
Scooped by
mhryu@live.com
January 27, 2:24 PM
|
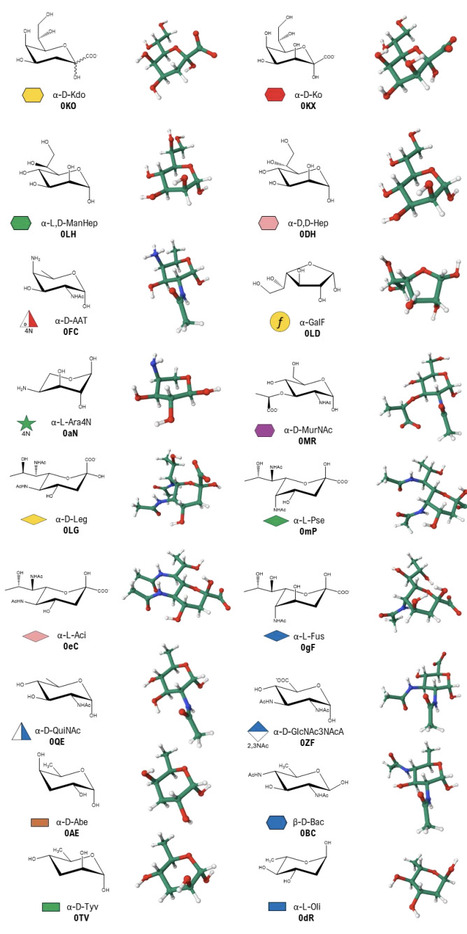
Here we present the GLYCAM Bacterial Carbohydrate Builder, an enhanced version of the GLYCAM-Web Carbohydrate Builder1 structure modeller that integrates support for modelling bacterial glycans, enabling the straightforward generation of three-dimensional structural models. The tool integrates bacterial monosaccharide parametrisations into a curated, user-friendly web-based resource. It provides an intuitive interface for the generation of carbohydrate sequences and generates 3D structural models in PDB file format, as well as the input files required for performing molecular dynamics simulations with the AMBER software package. The current implementation includes a library of 18 bacterial monosaccharides, which can be used in combination with the already parametrised eukaryotic sugars to construct complex bacterial glycans. Common derivatives, including acetylation, methylation, and sulfation are also supported. By validating and integrating bacterial sugar parameters into the GLYCAM-Web Carbohydrate Builder, this work reduces the technical barriers associated with bacterial glycan modelling and facilitates computational studies of complex bacterial glycoconjugates.

|
Scooped by
mhryu@live.com
January 27, 2:01 PM
|
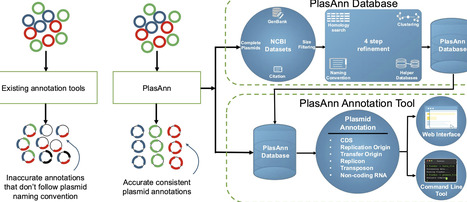
Conjugative plasmids are key drivers of bacterial adaptation, enabling the horizontal transfer of accessory genes within and across diverse microbial populations, yet annotating them remains challenging due to their highly mosaic genetic architectures and inconsistent gene naming conventions that complicate functional predictions and comparative analyses. To address this, we developed PlasAnn, a database designed specifically for genes encoded on natural plasmids, paired with a dedicated annotation pipeline (available via Bioconda or through the URL https://plasann.rochester.edu/). The curated database provides highly accurate, plasmid-type-specific gene names with standardized functional annotations, enabling direct comparison across plasmids without manual curation or specialized expertise, while the integrated annotation tool incorporates other common plasmid features for a fast, one-stop solution that outperforms broad prokaryotic genome annotation pipelines in both accuracy and efficiency. We demonstrate PlasAnn’s utility by showing that plasmid accessory genes from different groups often share conserved repertoires, suggesting dynamic, modular networks of interconnected genes, and by revealing that plasmid-encoded transposable elements frequently carry genes related to bacterial adaptation beyond antibiotic resistance, including metabolism, virulence, and stress responses, emphasizing their broader contributions to fitness and adaptability. These insights, not captured by current field-standard tools, highlight how PlasAnn improves plasmid annotation and advances our understanding of plasmid biology, microbial ecology, and evolution.

|
Scooped by
mhryu@live.com
January 27, 1:54 PM
|
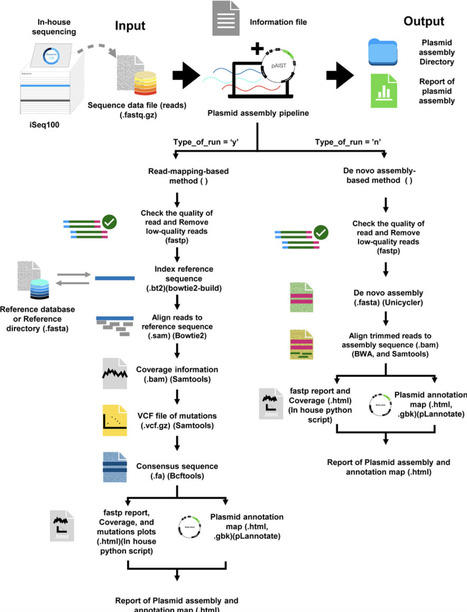
Sequence verification of plasmids is a fundamental process in synthetic biology. For plasmid sequence verification using next-generation sequencing (NGS) library preparation, Tn5 transposase is widely used. Streamlined sequencing workflow for laboratory-scale applications is important; however, recombinant Tn5 production in-house can be laborious. In this study, we demonstrated that the addition of a large soluble tag was not essential for purification and that the fusion of a His10 tag and protein A was sufficient to yield active Tn5 transposase in adequate amounts. In addition, we evaluated exonuclease-based genomic DNA digestion for plasmid sequencing from an E. coli lysate and the data analysis pipeline of sequences derived from the Illumina iSeq100 platform for de novo assembly, reference mapping, and annotation. This study proposes a simple workflow of an in-house easy-to-purify Tn5-based plasmid sequencing platform using a compact benchtop sequencer (AistSeq).

|
Scooped by
mhryu@live.com
January 27, 12:40 PM
|
Single-cell protein (SCP) is gaining attention as a source of food and feed, offering a lower environmental footprint than traditional agriculture. Efficient SCP production requires high cell density culturing (HCDC; >20 g cell dry weight L− 1), but O2 supply can become limiting in conventional aerobic systems. To circumvent this bottleneck, we recently proposed an anaerobic strategy using nitrate as an electron acceptor, exploiting denitrification-driven alkalinization in a pH-stat system to regulate substrate provision. Using the model organisms Paracoccus denitrificans and Paracoccus pantotrophus, we achieved biomass concentrations of up to 60 g dry weight L− 1 and protein contents up to 75% (75 ± 5%) in a 3 L fed-batch bioreactor. However, growth rates at high cell density were markedly lower than µmax observed in low-density cultures. In vivo and in silico experiments revealed three interacting constraints: (i) a CO2-pH lag where CO2 accumulation delayed alkalization by denitrification and thus substrate injection; (ii) mixing limitations, leading to poor substrate distribution; and (iii) physiological stress from nitrite accumulating during imbalanced denitrification. The CO2-pH lag emerged as the dominant barrier, resulting in long starvation periods. Lowering the pH setpoint of the pH-stat accelerated CO2 removal and thus substrate provision, but intensified nitrite toxicity. Insufficient mixing compounded the growth limitations as nitrate was only briefly available to a fraction of the population. Small-batch bioassays ruled out accumulation of inhibiting compounds other than nitrite. However, cells grown at high density in the reactor displayed reduced respiration rates, suggesting chronic stress under these conditions. Anaerobic HCDC by denitrification is feasible and yields high-quality biomass, but barriers remain to achieving competitive production rates. The CO2-pH lag appears to be the primary constraint, amplified by incomplete mixing and nitrite toxicity. These factors interact, e.g. mitigating the CO2-pH lag by lowering the process pH exacerbates nitrite toxicity. Future work should integrate reactor engineering to improve mixing and gas removal and strain selection for tolerance to low pH and nitrite, supported by omics and metabolic modelling to understand denitrifier physiology at high cell density.

|
Scooped by
mhryu@live.com
January 27, 10:44 AM
|
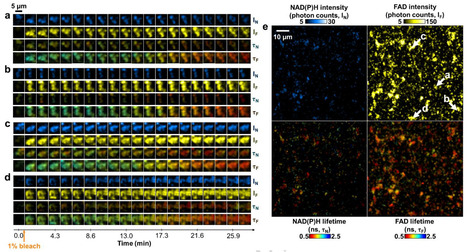
Label-free optical imaging provides non-invasive, high-speed, high-resolution metabolic characterization of live bacteria with single-cell resolution. Here, we demonstrate the ability of label-free multiphoton autofluorescence microscopy to characterize the fast (between 0 and 30 min) metabolic changes in bacteria in response to antibiotic treatments and observe the cell-to-cell metabolic heterogeneity of planktonic bacteria and biofilms. Results indicate that bacteria exhibit a distinct measurable response to bactericidal treatments within seconds. Furthermore, S. aureus biofilms exhibit metabolic heterogeneity, with local pockets of high metabolic activity. Bacteria in biofilms exhibit altered metabolic profiles compared to planktonic bacteria for all four species examined: S. aureus, P. aeruginosa, M. catarrhalis, and S. pneumoniae. These results shed light on the spatial and temporal metabolic heterogeneity of bacteria and the quantification possibilities using label-free nonlinear optical microscopy.

|
Scooped by
mhryu@live.com
January 27, 10:32 AM
|
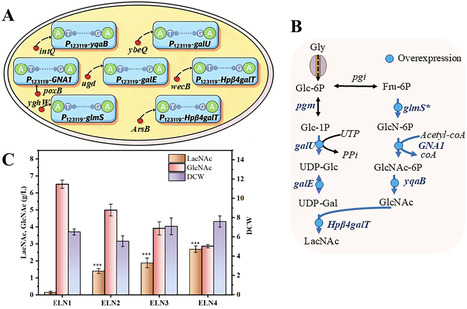
The biosynthetic production of human milk oligosaccharides through metabolic engineering has gained increasing attention in recent years. Yet, limited research has focused on the microbial synthesis of 3′-sialyllactosamine (3′-SLN), a critical intermediate for assembling sialyl Lewis X (sLex). In this work, a pathway enabling 3′-SLN biosynthesis was constructed in E. coli BL21(DE3). A key α2,3sialyltransferase (α2,3SiaT) from Bibersteinia trehalosi was identified and utilized to enhance the sialylation step toward 3′-SLN formation. To improve flux through the N-acetyllactosamine branch, essential genes were chromosomally integrated and overexpressed, while a CMP-N-acetylneuraminic acid synthesis module was coexpressed to secure an adequate donor supply. Furthermore, the expression balance was optimized through combinatorial adjustment of gene copy numbers and translational strength across modules. The engineered E. coli strain produced 0.78 g/L of 3′-SLN in shake-flask cultivation and further accumulated up to 7.75 g/L during 5-L fed-batch fermentation conditions, confirming the feasibility of microbial synthesis of 3′-SLN.
|
 Your new post is loading...
Your new post is loading...











Syn3A’s unusually high abundance of positively charged proteins is a crucial ingredient for chromosome expansion. These proteins enrich around ribosomes and DNA, partially charge-shielding ribosomes, which enables them to enter the nucleoid mesh and stabilize a cell-spanning nucleoid.