Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 5:01 PM
|
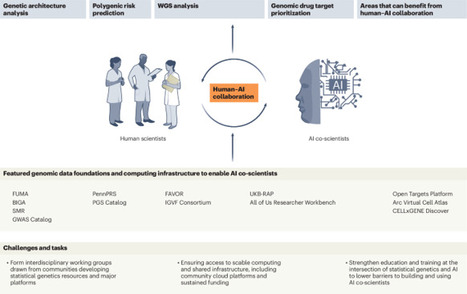
AI co-scientists can act as virtual research collaborators in statistical genetics, accelerating genetic discovery and translation. Realizing this potential depends on robust domain-specific data and infrastructures, together with interdisciplinary teamwork to build community standards that ensure rigor and responsible deployment.

|
Scooped by
mhryu@live.com
Today, 4:12 PM
|
Most genes in whose promotor a transcription factor (TF) binds do not change in expression when the concentration of the TF is perturbed. No existing model can predict which bound promotors will respond and which will not. We hypothesized that a gene's response to perturbation of a TF bound in its promotor can depend on which other TFs are bound there, a phenomenon we call functional synergy. This is distinct from cooperative binding, which is already accounted for in the binding location data. To investigate functional synergy, we created a comprehensive dataset on TF binding locations in yeast using a method that is orthogonal to chromatin immunoprecipitation. We then used mathematical modeling to identify high-confidence instances of functional synergy. We found that such synergies are surprisingly common. Responses to perturbations of 44 different TFs were modified by the presence of other TFs. 48 TFs served as modifiers, but some modified responses to many TFs. We conclude that (1) measuring the binding locations of a single TF will not, in general, reveal which genes the TF regulates, and (2) traditional networks linking TFs to their targets must be made substantially more expressive, allowing some TFs to modify the effects of others.

|
Scooped by
mhryu@live.com
Today, 2:01 PM
|
Global accumulation of difficult-to-recycle plastics, including polyurethane (PU), poses a significant environmental threat, as current recycling strategies are insufficient to meet the growing demand. Pseudomonas protegens is capable of degrading PU and may offer a promising nature-inspired solution. However, the genetic and regulatory mechanisms underlying microbial PU biodegradation remain poorly understood. To address this, we employed transposon insertion sequencing (TIS), a high-throughput functional genomics approach, to identify genes essential for PU degradation under plastic exposure conditions. A transposon mutant library of P. protegens Pf-5 was enriched on two types of PU: a polyester-polyurethane dispersion (Impranil DLN) and a thermoplastic polyurethane (Avalon AE). Through this approach, we identified a key global regulator encoding a sensor kinase, gacS, that plays an inhibitory role in PU degradation, so that Pf-5 lacking gacS displayed enhanced capacity to degrade PU. Transcriptomic and phenotypic analyses revealed that disruption of gacS resulted in increased activity of siderophore-mediated iron acquisition, while negatively impacting sulfur homeostasis and oxidative stress responses. We propose that this imbalance in iron homeostasis and oxidative stress in the gacS mutant triggers a Fenton reaction, leading to the accumulation of reactive oxygen species, which in turn enhances PU degradation via oxidative processes. These findings establish that siderophores involved in iron acquisition, such as pyoverdine, could act as catalysts for plastic degradation. Our results not only provide a deeper understanding of the genetic and regulatory basis of PU biodegradation but also open new avenues for leveraging bacterial pathways and siderophores for innovative plastic recycling strategies.

|
Scooped by
mhryu@live.com
Today, 1:54 PM
|
Assigning mechanistic and functional associations to most of the widespread splicing diversity in fungi has been one of the continuous and most intriguing challenges in the field of molecular mycology. Ongoing advances in sequencing technologies have driven recent outbreaks in the study of the dynamic splicing landscapes of fungal transcriptomes. Nevertheless, most ascomycete and basidiomycete species of agronomic, industrial, and medical interest exhibit a notable proportion of their transcripts affected by alternative splicing (AS), producing mRNA isoforms whose regulation and functions have been largely ignored to date. In contrast to all other fungi, some yeast and basal fungal clades exhibit extremely scarce AS occurrences, which are associated with very low genomic intron counts. In this review, we provide thoughtful insights into three long-standing unsolved questions: (1) the pervasive occurrence and possible stochasticity of intron retention in fungi, (2) the reasons for increased AS rates in pathogenic and filamentous species, and (3) possible hypotheses to explain the functionality and fitness advantages of AS in fungi. We present a critical discussion on the role of AS, highlighting its importance in an evolutionary context that integrates major features of fungal diversification and adaptability. To advance our understanding of fungal splicing control, function, and evolution, we propose a practical roadmap for future research, aimed at filling the most critical gaps in the field.

|
Scooped by
mhryu@live.com
Today, 1:40 PM
|
TnPedia provides a curated framework for understanding prokaryotic transposable elements (TEs), including general and historical information and basic concepts, while integrating mechanistic insights, family-level classification, and representative examples. It summarizes key developments reshaping the field, including RNA-guided transposition, and provides information on TE acquisition and transmission of antimicrobial resistance-associated transposons.

|
Scooped by
mhryu@live.com
Today, 1:36 PM
|
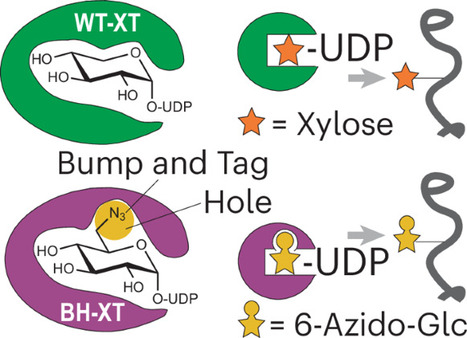
Mammalian cells receive signaling instructions through interactions on their surfaces. Proteoglycans are critical to these interactions, carrying long glycosaminoglycans that recruit signaling molecules. Biosynthetic redundancy in the first glycosylation step by two xylosyltransferases XT1/2 complicates annotation of proteoglycans. Here we develop a chemical genetic strategy that manipulates the glycan attachment site of cellular proteoglycans. Through a bump-and-hole tactic, we engineer the two isoenzymes XT1 and XT2 to specifically transfer the chemically tagged xylose analog 6AzGlc to target proteins. The tag contains a bioorthogonal functionality, allowing to visualize and profile target proteins in mammalian cells. Unlike xylose analogs, 6AzGlc is amenable to cellular nucleotide-sugar biosynthesis, establishing the XT1/2 bump-and-hole tactic in cells. The approach allows pinpointing glycosylation sites by mass spectrometry and exploiting the chemical handle to manufacture proteoglycans with defined glycosaminoglycan chains for cellular applications. Engineered XT enzymes permit an orthogonal view into proteoglycan biology through conventional techniques in biochemistry. The xylosyltransferase isoenzymes XT1 and XT2 catalyze the first glycosylation step in the biosynthesis of proteoglycans. Now, bump-and-hole engineering of XT1 and XT2 enables substrate profiling and modification of proteins as designer proteoglycans to modulate cellular behavior.

|
Scooped by
mhryu@live.com
Today, 1:16 PM
|
Agricultural sustainability faces serious challenges from population growth, climate change and ecological degradation. Genetic modification (GM) technology can be regarded as a precise extension of the Green Revolution, aiming to balance yield enhancement with ecological integrity through biotechnology. To systematically examine global trend, this study conducts a bibliometric analysis using worldwide literature data from 1994 to 2024. The findings reveal a dual-core structure of international collaboration, centered on China and the United States. The United States is closely connected with Korea, Japan and the United Kingdom, forming a high-density cluster, while China engages with emerging regions in Southeast Asia and Africa through the Belt and Road Initiative. This initiative is intended to strengthen China's influence and is accompanied by the proliferation of technology in countries less endowed with resources. The technology lifecycle has been evolved through three distinct phases. Initially, the process of Agrobacterium-mediated transformation in tobacco plants was carried out, marking the beginning of transgenic development. This was followed by the implementation of RNA interference (RNAi) technology to silence multiple genes. Finally, a breakthrough happened through the development of CRISPR-Cas9 genome editing technologies. The analyses conducted in this study demonstrate the preponderance of CRISPR in contemporary research, thus suggesting that the industry places a premium on technological refinement. Hence, the future technological trajectory is predicted to focus on germplasm digitization, multi-gene editing, intelligent breeding and synthetic biology. Transgenic technology will serve as a foundational support for achieving sustainable food security in the forthcoming second green revolution.

|
Scooped by
mhryu@live.com
Today, 1:02 PM
|
Transcription factor structural dynamics has emerged as a critical frontier in plant molecular biology, as many transcription factors (TFs) belong to plant-specific families with unique structural features that reveal mechanisms static structural approaches cannot capture. These advances, combining molecular dynamics simulations, NMR spectroscopy, and single-molecule techniques, have demonstrated that structural dynamics play crucial roles in driving DNA recognition, environmental responsiveness, and regulatory precision. Such computational and experimental breakthroughs have revealed rotation-coupled diffusion processes in WRKY proteins and allosteric regulation through ensemble redistribution in disordered domains. Despite these discoveries, fundamental questions remain about how plant TFs achieve specificity within complex regulatory networks and integrate multiple environmental signals. We outline current understanding of the static structures across transcription factor families before examining breakthrough discoveries in structural dynamics. These encompass the diverse architectures of DNA binding and regulatory domains, as well as the conformational flexibility and transition kinetics governing DNA binding, cofactor interactions, and environmental signal transduction. We subsequently explore how these insights are transforming biotechnological applications, from rational protein design to crop engineering strategies. Finally, we discuss remaining challenges and future directions for harnessing transcription factor dynamics in agricultural and synthetic biological applications.

|
Scooped by
mhryu@live.com
Today, 12:51 PM
|
This study explores how Pseudomonas putida EM2-4 metabolizes linear alkanes after acquiring an integrative and conjugative element (ICE) encoding a novel alkane monooxygenase (AlkB). This AlkB enzyme is a fusion of a hydroxylase and a fatty acid desaturase, exhibiting a narrow substrate range limited to n-octane and n-decane. The oxidation of these hydrocarbons yields octanoate and decanoate, respectively, which then enter the β-oxidation pathway and the glyoxylate shunt. Under nitrogen-limiting conditions, excess carbon is redirected toward polyhydroxyalkanoate (PHA) synthesis, a phenomenon particularly pronounced in a phaZ-deficient mutant unable to depolymeraze PHA, leading to up 75% of cell dry weight. Analysis of the PHAs monomer composition revealed variations with the carbon source. Additionally, 13C-isotopic labeling uncovered a minor but unexpected fraction of C10 monomers from C8 substrates, potentially arised from gluconeogenesis followed by the de novo synthesis of fatty acids. Proteomic analyses showed that the carbon source influences the activation of metabolic pathways, correlating with the observed variations in PHAs monomer composition.

|
Scooped by
mhryu@live.com
Today, 12:42 PM
|
Bacteriophages have evolved diverse strategies to manipulate host processes, yet the molecular mechanisms employed by phage-encoded effector proteins remain poorly understood. Here, we identify Gp16, an early-expressed protein from Staphylococcus aureus phage ΦNM1, as a RecA-dependent nuclease that plays a dual role in host inhibition and phage propagation. Gp16 is rapidly expressed upon infection, and its overexpression alone is sufficient to inhibit bacterial growth where deletion of gp16 severely impairs phage DNA replication, progeny production, and host cell lysis, underscoring its essential role in the phage life cycle. Structural modeling predicts Gp16 is a nuclease, and its overexpression induces DNA condensation in vivo. Biochemical and cellular analyses show that Gp16 interacts with the host RecA protein to inhibit growth, and functions as a nickase in vitro, requiring the catalytic cysteine C181 for DNA cleavage. RecA further enhances its cleavage activity. During phage infection, RecA activation is required for efficient phage propagation, while Gp16 concurrently suppresses host DNA replication and promotes DNA condensation, thereby facilitating phage replication. Together, these findings reveal a previously unrecognized strategy in which a phage-encoded nuclease exploits the host RecA machinery to couple host suppression with productive phage propagation.

|
Scooped by
mhryu@live.com
Today, 12:29 PM
|
Root-feeding herbivores impose substantial constraints on plant performance, yet the chemical signals coordinating belowground defence remain poorly resolved. Microbial volatile organic compounds (VOC)—highly diffusible metabolites produced by rhizosphere and endophytic microbes—have emerged as pivotal regulators of plant immunity and development. Recent evidence shows that specific mVOCs modulate jasmonic acid, salicylic acid, ethylene, auxin, and ROS-associated pathways, thereby reprogramming root architecture and priming defence responses during herbivore attack. Despite these advances, major mechanistic gaps persist, including how plants perceive mVOCs, how soil physicochemical conditions shape their diffusion and bioactivity, and how mVOCs integrate with plant-derived volatiles and metabolites to coordinate systemic signalling. Moreover, the roles of mVOCs in mediating multitrophic interactions—particularly their influence on root herbivore behavior, microbial recruitment, and defence hormone crosstalk—remain largely unexplored. This review synthesizes current advances in mVOC biology and proposes conceptual frameworks linking microbial volatilomes to plant hormonal networks and belowground herbivore defence. A deeper understanding of these hidden chemical dialogues will inform strategies for enhancing crop resilience and developing sustainable root pest management.

|
Scooped by
mhryu@live.com
Today, 10:30 AM
|
Microbes are powerful systems for exploring and engineering biology. Their compact genomes, rapid generation times, and experimental tractability enable quantitative analyses that shed light on both conserved cellular mechanisms and traits of medical or industrial relevance. Building on this foundation, systematic perturbation has become central to microbial systems biology. Genome-scale knockout and CRISPRi/a libraries have mapped gene function and network architecture, yet these approaches largely operate at the level of gene presence or absence, leaving the effects of precise sequence variants unexplored. Recent and emerging precision-perturbation strategies now reveal biological principles inaccessible to gene-level perturbations, from detailed sequence–function maps of proteins to the impact of natural and engineered variation across pathways. In this review, we highlight recent advances that have made systematic interrogation of thousands of variants — within single loci and across entire genomes — increasingly comprehensive and efficient. We will discuss how these technical leaps reveal systems-level principles of genome function and provide outlooks on how they could be complemented by diverse phenotypic readouts and perturbations in combinatorial space. Taken together, empowering precision engineering approaches will further advance our understanding of biological function, while accelerating progress in biotechnology and synthetic biology.

|
Scooped by
mhryu@live.com
Today, 12:49 AM
|
Eukaryotic Fanzor proteins are compact, programmable RNA-guided nucleases with substantial potential for genome editing, although their efficiency in mammalian cells remains suboptimal. Here, we present a combinatorial engineering strategy to optimize a representative Fanzor system, MmeFz2–ωRNA. AlphaFold3-powered rational redesign produced a minimized ωRNA scaffold that is 30% smaller while maintaining up to 82.2% efficiency. Synergistic structure-guided and AI-augmented protein engineering generated two variants, enMmeFz2 and evoMmeFz2, which exhibited an average ~32-fold increase in activity across 38 genomic loci. Moreover, fusion of the non-specific DNA-binding domain HMG-D further enhanced editing performance (enMmeFz2-HMG-D and evoMmeFz2-HMG-D). Notably, evoMmeFz2-HMG-D demonstrated robust in vivo genome editing activity, enabling dystrophin restoration in humanized male Duchenne muscular dystrophy mouse models via single adeno-associated virus (AAV) delivery. This study establishes Fanzor2 as a gene editing platform for genome engineering and therapeutic applications, and underscores the power of AI-guided engineering to accelerate genome editor development while reducing experimental burden. Eukaryotic Fanzor proteins are compact and advantageous for delivery, but their activity remains limited. Here, the authors engineer an improved Fanzor2 system (evoMmeFz2) using structure-guided and AI-assisted strategies to enable efficient exon skipping in a Duchenne muscular dystrophy model.
|

|
Scooped by
mhryu@live.com
Today, 4:22 PM
|
Microcins are small antibacterial proteins secreted by gram-negative bacteria. The activities of new microcins discovered using bioinformatic searches need to be validated and characterized to understand how they mediate competition in microbiomes and to evaluate their potential as new therapeutics for combating antibiotic resistance. Engineered plasmids containing the type I secretion system associated with E. coli Microcin V (MccV) can secrete heterologous proteins, including other class II microcins, and this system functions in other bacterial hosts. However, existing microcin secretion constructs are not designed for easily swapping components — such as origins of replication, resistance genes, promoters, and signal peptides — that may need to be changed for compatibility with other chassis. We refactored the E. coli MccV type I secretion system into genetic parts compatible with a modular Golden Gate assembly scheme and used these parts to construct two-plasmid microcin secretion systems. In our design, one plasmid encodes the type I secretion system proteins, and the other encodes a signal peptide fused to the cargo protein that will be secreted. We tested two versions of a system with inducible promoters separately controlling expression of the components on each plasmid. One used plasmids that replicate in E. coli and its close relatives. The other used broad-host-range plasmids. When induced to secrete MccV, both versions produced similar zones of inhibition against a susceptible strain of E. coli. Next, we identified putative class II microcins in genomes of bacteria from plant-associated genera (Pantoea, Erwinia, and Xanthomonas) using an existing bioinformatics pipeline. We screened 23 of these putative microcins for E. coli self-inhibition. Seven exhibited some inhibition, mostly later in growth curves, but none had effects that were comparable in strength to MccV. The genetic parts we created can be assembled in various combinations into tailored systems for secreting small proteins from diverse bacterial chassis. These systems can be used to further characterize the targets of novel microcins and to secrete these or other small proteins for various applications. For example, beneficial bacteria used in crop protection could be engineered to secrete microcins that kill or inhibit plant pathogens to increase their efficacy.

|
Scooped by
mhryu@live.com
Today, 3:52 PM
|
Continuous cropping often exacerbates soil-borne diseases, particularly Fusarium wilt, yet the intricate rhizosphere relationships among phyto-derived metabolites, pathogens, and particular microbial functions remain poorly understood. Here, we observe that citrulline accumulation during continuous cropping is positively correlated with Fusarium wilt severity by enhancing fusaric acid production in Fusarium oxysporum. Metagenomic analyses reveal that citrulline turnover-related functions, represented by functional modules including M00978, are significantly enriched in healthy rhizosphere soils but are notably reduced in Fusarium-conducive soils. The functional genes, arcB and argH, are identified in Pseudomonas putida YDTA3, with arcB being essential for citrulline-degradation via knockout experiments. The inoculation of an arcB-expressing indigenous Escherichia consortium (EO-arcB) in three independent continuous cropping systems of cucurbit crops demonstrates that enhancing and maintaining the soil citrulline-degrading function mitigates soil-borne Fusarium wilt. In summary, this study advances our understanding of rhizosphere interactions underlying Fusarium wilt disease occurrence and offers a promising biocontrol strategy. In this study, the authors show that citrulline accumulation exacerbates Fusarium wilt and that enhancing the soil’s citrulline-degrading function, particularly via the arcB gene, significantly reduces disease in continuous cucurbitaceous cropping systems.

|
Scooped by
mhryu@live.com
Today, 1:56 PM
|
Bacteriophages are considered to have great potential as naturally occurring, antimicrobial agents for use in food production. Phages are ubiquitous in nature and can be isolated from almost all habitats. This review outlines the possibilities, as well as limitations of their use in food production. Application of phages in the food sector are described and the limitations of their use as well as potential risks are discussed. Approaches for a possible classification as either processing aid or food additive are considered, and the current status of their use in and outside the EU is presented. Finally, the need for research to close identified knowledge gaps is highlighted.

|
Scooped by
mhryu@live.com
Today, 1:46 PM
|
Coinfection of Pseudomonas aeruginosa and Staphylococcus aureus is frequently observed. Our previous study demonstrated that S. aureus-derived extracellular vesicles (SaEVs) promote P. aeruginosa pathogenicity by increasing lipopolysaccharide (LPS) production, promoting biofilm formation and decreasing the uptake of P. aeruginosa by macrophages. Proteomic analysis revealed that SaEVs enhance the production of PslE, an exopolysaccharide biosynthetic protein in P. aeruginosa, but the role of Psl exopolysaccharide polymerization on SaEV-mediated P. aeruginosa pathogenicity is unclear. In this study, a pslE-deletion mutant of P. aeruginosa (PaΔpslE) was constructed, and the effect of SaEVs on the pathogenicity of this mutant was evaluated. Our results showed that SaEVs significantly increased the expression of pslA, E, J, K, and L genes in the psl cluster of P. aeruginosa wild-type (PaWT), and this effect was abolished in PaΔpslE. In addition, LPS production and biofilm formation were reduced in PaΔpslE compared to PaWT. SaEVs significantly enhanced LPS production and biofilm formation in PaWT. On the other hand, the effects of SaEVs on the production of lipid A and LPS core and biofilm formation in PaΔpslE were abolished. Invasion of PaWT and PaΔpslE into HaCaT human epithelial cells was not significantly different and the effect of SaEVs on these bacterial cell invasions was not found. However, the uptake of SaEV-treated PaWT by macrophages significantly reduced compared to nontreated PaWT, whereas SaEVs did not alter the uptake of PaΔpslE. These results suggest that PslE is required for SaEV-mediated P. aeruginosa pathogenicity. SaEVs upregulate pslE gene as well as other exopolysaccharide polymerization-related genes, increase LPS production and biofilm formation, and affect the uptake of P. aeruginosa by macrophages.

|
Scooped by
mhryu@live.com
Today, 1:39 PM
|
This review covers the literature published in 2024 for marine natural products (MNPs), with 617 citations (578 for the period January to December 2024) referring to compounds isolated from marine microorganisms and phytoplankton, green, brown and red algae, sponges, cnidarians, bryozoans, molluscs, tunicates, echinoderms, the submerged parts of mangroves and other intertidal plants. The emphasis is on new compounds (1256 in 336 papers for 2024), together with the relevant biological activities, source organisms and country of origin. Pertinent reviews, biosynthetic studies, first syntheses, and syntheses that led to the revision of structures or stereochemistries, have been included. An analysis of the role of artificial intelligence in marine natural products research is discussed.

|
Scooped by
mhryu@live.com
Today, 1:31 PM
|
With the increasing demand for sustainable agriculture, rhamnolipids (RHAs), a class of bio-surfactants, have emerged as promising eco-friendly biostimulants. However, their full potential has been limited by impurities and uncertain mechanisms. This study investigated the effects of high-purity RHAs on wheat (Triticum aestivum) growth and yield through controlled laboratory experiments and multiyear field trials. In laboratory assays, optimal RHA concentrations (5–10 mg·L–1) promoted seedling growth by enhancing chlorophyll content, root–shoot development, and tillering. Transcriptomic profiling revealed that RHA treatment upregulated genes involved in gibberellic acid (GA3) and zeatin (ZT) biosynthesis, nitrogen uptake, and photosynthesis. In the field, low doses of RHA application (∼45 g·ha–1) significantly enhanced tillering capacity and promoted earlier flowering while delaying senescence. Importantly, these physiological enhancements translated into significant yield increases of 11.9 and 14.8% across 2 consecutive years. The study provides novel mechanistic insights into RHA-mediated growth promotion, highlighting RHAs as a promising, eco-friendly biostimulant for sustainable wheat production.

|
Scooped by
mhryu@live.com
Today, 1:03 PM
|
Fiber, the most abundant organic polymer in nature, is widely recognized as a foundational sustainable material with diverse applications across industrial, medical, and consumer domains. Owing to its renewability and widespread availability, it also serves as a critical alternative energy source in agriculture, enabling more sustainable livestock production through the efficient conversion of fibrous feedstuffs, thereby supporting the principles of a circular bioeconomy. Cellulose, which constitutes up to 80% of plant fiber, contains tightly packed crystalline regions that confer strong resistance to microbial degradation. Other key obstacles to efficient cellulose digestion in the gut include the absence of critical cellulolytic genes, low enzymatic activity, a lack of natural activators, and the presence of cellulase inhibitors. Synthetic biology provides innovative molecular-level strategies to overcome key technical barriers in cellulose degradation. These approaches employ targeted modifications at nucleic acid and protein levels, including the introduction of engineered genes, synthetic regulators, and optimized enzymes, to develop high-performance microbial systems with enhanced cellulose-degrading capabilities. Furthermore, genetic modifications like the knockout of inhibitory genes and knock-in of activator genes, combined with rational redesign of multi-enzyme complexes, can significantly improve the secretion and catalytic efficiency of cellulases. When integrated with artificial intelligence, synthetic biology enables predictive screening and precision engineering of microbial strains for highly efficient cellulose degradation. This review comprehensively summarizes recent advances in synthetic biology approaches for improving cellulose degradation and highlights how these tools can optimize fiber utilization in sustainable agricultural and industrial applications.

|
Scooped by
mhryu@live.com
Today, 12:54 PM
|
The nitrogen-fixing symbiosis between leguminous plants and soil bacteria, collectively termed rhizobia, is a major contributor of fixed nitrogen to the biosphere. The ability of legumes to secure nitrogen from the atmosphere underlies their ecological success and has made them important crops in both traditional and modern sustainable agriculture. Many genes directing the establishment and functioning of this beneficial interaction have been identified under laboratory conditions using a limited number of bacterial strains and plant species in pairwise combinations. Under natural and field conditions, however, plants encounter numerous potential partners, as soil microbiomes contain diverse bacteria equipped with the necessary toolkit for symbiosis. Consequently, legumes must possess mechanisms to select for or against specific partners. This review highlights how legumes employ elements of their immune system for the negative selection of rhizobia via processes resembling the gene-for-gene model of effector-triggered immunity in plant–pathogen interactions.

|
Scooped by
mhryu@live.com
Today, 12:44 PM
|
TenA (transcription enhancement) family proteins are widespread in bacteria, yet their functions in regulating natural product biosynthesis remain largely unexplored. Here, we report that LysR2, a TenA-family protein encoded by the lysolipin I biosynthetic gene cluster, acts as a transcriptional activator. Genetic and biochemical analyses reveal that LysR2 employs a noncanonical mechanism. Lacking a standard DNA-binding domain, it does not bind target promoters directly. Instead, LysR2 activates transcription by specifically binding to the pathway-specific repressor, LysR1. This protein–protein interaction displaces LysR1 from its cognate promoter, thereby derepressing the transcription of the biosynthetic gene cluster. Our work provides the first experimental evidence for a regulatory role of a TenA-family protein and elucidates a unique indirect activation mechanism that broadens the paradigm of transcriptional regulation in bacterial natural product biosynthesis.

|
Scooped by
mhryu@live.com
Today, 12:36 PM
|
Knowledge graphs (KGs) are powerful tools for structuring and analyzing biological information due to their ability to represent data and improve queries across heterogeneous datasets. However, constructing KGs from unstructured literature remains challenging due to the cost and expertise required for manual curation. Prior works have explored text-mining techniques to automate this process, but have limitations that impact their ability to capture complex relationships fully. Traditional text-mining methods struggle with understanding context across sentences. Additionally, these methods lack expert-level background knowledge, making it difficult to infer relationships that require awareness of concepts indirectly described in the text. Large Language Models (LLMs) present an opportunity to overcome these challenges. LLMs are trained on diverse literature, equipping them with contextual knowledge that enables more accurate information extraction. We present textToKnowledgeGraph, an artificial intelligence tool using LLMs to extract interactions from individual publications directly in Biological Expression Language (BEL). BEL was chosen for its compact, detailed representation of biological relationships, enabling structured, computationally accessible encoding. This work makes several contributions. 1. Development of the open-source Python textToKnowledgeGraph package (pypi.org/project/texttoknowledgegraph) for BEL extraction from scientific articles, usable from the command line and within other projects, 2. An interactive application within Cytoscape Web to simplify extraction and exploration, 3. A dataset of extractions that have been both computationally and manually reviewed to support future fine-tuning efforts.

|
Scooped by
mhryu@live.com
Today, 10:37 AM
|
Following their success in natural language processing and protein biology, pretrained large language models have started appearing in genomics in large numbers. These genomic language models (gLMs), trained on diverse DNA and RNA sequences, promise improved performance on a variety of downstream prediction and understanding tasks. In this review, we trace the rapid evolution of gLMs, analyze current trends, and offer an overview of their application in genomic research. We investigate each gLM component in detail, from training data curation to the architecture, and highlight the present trends of increasing model complexity. We review major benchmarking efforts, suggesting that no single model dominates, and that task-specific design and pretraining data often outweigh general model scale or architecture. In addition, we discuss requirements for making gLMs practically useful for genomic research. While several applications, ranging from genome annotation to DNA sequence generation, showcase the potential of gLMs, their use highlights gaps and pitfalls that remain unresolved. This guide aims to equip researchers with a grounded understanding of gLM capabilities, limitations, and best practices for their effective use in genomics.

|
Scooped by
mhryu@live.com
Today, 9:30 AM
|
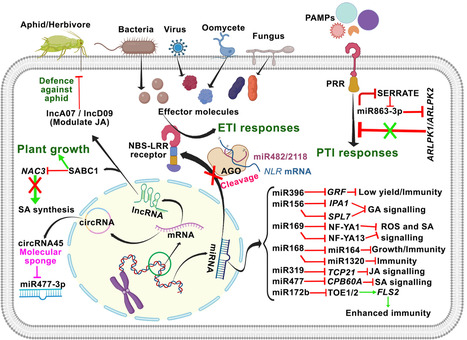
Balancing growth and immunity pose a fundamental challenge for plants, as investment in defence mechanisms often comes at the cost of growth and development, and vice versa. Ecological theories, particularly those based on principles of economic optimality, propose that plants strategically allocate resources between these competing processes to optimize fitness. Recent advances in multi-omics technologies have provided deep insights into the complex molecular architecture governing growth-defence trade-offs, identifying transcriptional cascades as key regulators of resource allocation. Within this regulatory landscape, non-coding RNAs (ncRNAs) have emerged as critical modulators, fine-tuning the expression of genes involved in both developmental and immune pathways. This review synthesizes current understanding of how ncRNAs mediate the growth-defence balance in plants. We discuss specific ncRNAs that act as regulatory hubs within feedback circuits, their interactions with phytohormonal signalling networks, and the potential of harnessing these mechanisms for precision crop improvement. Ultimately, decoding the functional framework of ncRNA-driven regulation presents new opportunities to develop high-yielding, stress-resilient crop varieties that can bypass the constraints of conventional growth-defence trade-offs, thereby supporting sustainable agricultural productivity amid growing environmental pressures.
|






 Your new post is loading...
Your new post is loading...