 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 12:45 PM
|
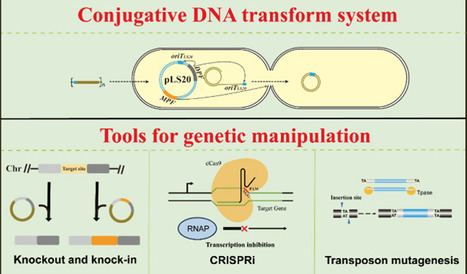
Bacillus licheniformis is a spore-forming bacterium with probiotic, environmental, and industrial applications. Many wild strains with diverse functions have been described in recent years. Nevertheless, the lack of efficient and universal genetic manipulation tools hinders the study and engineering of these strains. Here, a versatile and simple genetic manipulation toolkit is established for B. licheniformis. The cornerstone of this toolkit is a conjugative DNA transfer system. This system could effectively transfer temperature-sensitive plasmid pTSMK into all ten tested B. licheniformis strains, with efficiencies ranging from 10–5 to 10–3. Based on this DNA transfer system, the tools for maker-free knockout and knock-in, CRISPRi, as well as transposon mutagenesis, were built. A transposition frequency of 7.68 × 10–3 was observed. The toolkit developed in this study fulfills most tasks in the engineering of this species and will promote the basic and applied research of B. licheniformis.

|
Scooped by
mhryu@live.com
Today, 12:34 PM
|
Phytohormones are compounds produced by plants that can control the processes of plant development and are involved in defense mechanisms. They can serve as sustainable enhancers for plant growth and are also better alternatives to agrochemicals because of their bio-based nature and optimal safety profile towards mammalian cells. Although phytohormones are essential for plant growth, they are naturally synthesized by plants in varying quantities, influenced both by environmental and genetic factors. The synthesis of phytohormones, however, faces several hurdles, including challenges in optimizing the production process as well as difficulties in the extraction of these compounds for downstream applications. In this review, we critically evaluate synthetic biology approaches for phytohormone production, which provides a sustainable method for the production of important phytohormones including auxin, gibberellin (GA), and salicylic acid (SA). These three compounds are highly significant for the agricultural industry because of their widespread utilization.

|
Scooped by
mhryu@live.com
Today, 12:23 PM
|
Lignocellulosic biomass is the most abundant and sustainable carbon source for bioproduction, but its efficient utilization is hampered by the heterogeneous mixture of sugars released upon hydrolysis. Most industrial strains consume these mixed sugars sequentially due to strong regulation and cross-inhibition, leading to complex processes and reduced carbon efficiency. To address this, we leverage a novel central metabolism that decouples central carbon flux from native regulatory feedback by employing a Gluconate-Bypass of glycolysis. We demonstrate that the Gluconate-Bypass effectively alleviates feedback regulation in E. coli, enabling co-consumption of glucose and xylose. Further strain engineering leads to the first robust co-utilization of four major lignocellulosic sugars: glucose, xylose, arabinose, and galactose. By decoupling central carbon flux from native regulatory feedback, this architecture provides a feedstock-agnostic platform that maintains high and robust consumption regardless of extreme fluctuations in sugar composition.

|
Scooped by
mhryu@live.com
December 27, 9:59 PM
|
Protein-glutaminase (PG), as a novel food additive with a remarkable deamidation capacity, is naturally expressed as a proenzyme. Activation of the PG proenzyme is mostly dependent on proteases. To address this problem, an intein-mediated activation (IMA) system was developed for PG production using self-splicing inteins that can catalyze a single C-terminal cleavage. In this study, fusion proenzymes were expressed in Escherichia coli by inserting different inteins between the pro-peptide and mature PG and the C-terminal activation efficiency was evaluated in vitro. Subsequently, Mth RIR1 (RIR1) and Mxe GyrA (Mxe) were selected for construction of the IMA system in various Bacillus subtilis strains. Finally, the PG activity increased to 37.2 U/mL after promoter optimization and fed-batch fermentation in B. subtilis WB600. This approach enables one-step and nonprotease-dependent PG production in B. subtilis and establishing an alternative strategy for other proenzyme activation.

|
Scooped by
mhryu@live.com
December 27, 9:10 PM
|
Diatoms are key contributors to global primary production, and have developed intricate partnerships with bacteria through long-term co-evolution. Here, we uncover a syntrophic relationship between the model obligate photoautotroph diatom Phaeodactylum tricornutum and the rod-shaped bacterium Loktanella vestfoldensis, which enables the diatom to indirectly utilize glucose. To be specific, growth of the diatom depends on the support of L. vestfoldensis for the supply of necessary carbon source when glucose serves as the sole carbon source, while L. vestfoldensis shows dependence on P. tricornutum when CO2 is the sole carbon source. Reanalysis of Tara Oceans metagenomic data shows frequent co-occurrence of Loktanella with diatoms including Chaetoceros and Thalassiosira, indicating the ecological relevance of this partnership. Co-culture with L. vestfoldensis supports robust growth of Chaetoceros muelleri and Thalassiosira pseudonana in the presence of glucose as the sole carbon source. Transcriptomic and metabolomic analyses reveal that P. tricornutum maintains a photoautotrophic metabolism in co-culture, as indicated by the up-regulation of genes involved in inorganic carbon concentration and photosynthesis, while the co-cultured bacterium likely supplies CO2 and growth-stimulating metabolites such as indole-3-acetic acid. Our findings demonstrate that bacterial-algal interactions may shape diatom adaptation to carbon changes and contribute to marine carbon cycling. Diatoms are key marine primary producers and establish intricate partnerships with bacteria. Bacterium Loktanella enables diatoms to indirectly utilize glucose and maintains their photoautotrophic metabolism, indicating a common oceanic association.

|
Scooped by
mhryu@live.com
December 27, 12:29 PM
|
Bacteria-Fungi Interactions play a crucial role in soil nutrient cycling and plant disease suppression. Bacillus and Trichoderma exhibit antagonism when inoculated on laboratory media, global soil sample analysis reveals a positive correlation between these two genera in addition to enhanced plant-pathogen Fusarium oxysporum suppression and plant growth promotion. Here, we assess cross-kingdom interactions within artificial model communities of Bacillus velezensis and Trichoderma guizhouense. Transcriptomic profiling revealed that in the presence of fungi, the key stress sigma factor of B. velezensis activates expression of biosynthetic genes for antimicrobial secondary metabolite production. Among these, surfactin induces T22azaphilone production in T. guizhouense that hinders oxidative stress. Both surfactin and T22azaphilone contribute to Bacillus and Trichoderma maintenance in soil in the presence of Fusarium oxysporum. Finally, Fusarium oxysporum-secreted fusaric acid temporarily inhibits B. velezensis growth whereas it is efficiently degraded by T. guizhouense. These metabolite-mediated interactions reveal how competing soil microorganisms could form effective alliances that ultimately enhance plant protection against soil-borne pathogens.

|
Scooped by
mhryu@live.com
December 27, 12:09 PM
|
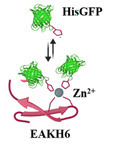
The development of tunable systems for subcutaneous injection is currently the focus of exploratory protein formulation. For the delivery of protein biologics intended for extended release, a high fraction of the drug that escapes from the deposition site (“burst release”) may pose safety concerns. Herein, we report an injectable system of bioaffinity zinc-containing peptide coacervates in which recombinant proteins coexpressed with histidine (His)-tags can be captured and released over time. Coacervates are formed by driving self-assembling peptide (SAP), AEAEAKAKAEAEAKAKHHHHHH (EAKH6) β-sheet dimers, into cross-linking fibrils. In the presence of Zn2+, the fibrillization of EAKH6 is enhanced through the interaction of metal ions with histidine residues in the peptide. The Zn2+:EAKH6 scaffold retains His-tagged proteins both in vitro and in vivo and extends their duration of release. The results present a case study in which the Zn2+–[His]6 interaction can be used to tune the properties of supramolecular structures and the loading of His-tagged proteins.

|
Scooped by
mhryu@live.com
December 27, 11:55 AM
|
The advent of synthetic biology, enabling the construction of synthetic genetic circuitry with designed functionality, has has a revolutionary impact on medicine, agriculture, sustainable energy, and the industrial production of high-value compounds over the last few decades. Gene switches have an indispensable role as regulators of such systems. Despite the early introduction of chemically inducible switches to regulate genetic circuitry, ‘traceless’ physical cues (e.g., light, heat, sound, magnetism, electricity, and mechanical force) can provide greater specificity, higher spatiotemporal resolution, more flexible switching patterns, and better compatibility with bioelectronic interfaces, which is of particular significance given the rise of electrogenetics. Indeed, traceless gene switches are on a path to become universal biological control ports interfacing physiology with the electronic world. In this review, we discuss the impact, challenges, and prospects of physically inducible, traceless gene switches in the context of recent cutting-edge applications.

|
Scooped by
mhryu@live.com
December 27, 11:34 AM
|
Continuous two-stage E. coli fermentations offer potential for high-efficiency bioprocesses but are often limited by plasmid instability, genetic mutations, and unintended expression during seed phases. In this study, we aimed to overcome these limitations by employing a plasmid-dependent auxotrophic selection system, specifically using a thymidine auxotrophy (thyA deletion), in conjunction with a series of plasmid modifications to enhance stability and expression control. We engineered the E. coli strain enGenes-eXpress V2 ΔthyA and constructed modified plasmids containing thyA along with regulatory elements to maintain plasmid stability and minimize basal expression. The modified strains were evaluated in continuous two-stage fermentations under carbon-limited conditions. Our results indicate significant reduction in plasmid loss, improved population homogeneity, and suppressed basal expression in non-induced phases. The addition of elements such as the cer (ColE1 resolution) site and a modified T7 promoter further enhanced plasmid stability and reduced basal expression levels. High-throughput screenings in microbioreactor setups confirmed that optimized constructs maintained a homogeneous producing population and suppressed non-producing cells over extended periods, which was validated by fed-batch cultivations and single-cell analyses. Finally, the two most promising constructs demonstrated high robustness in continuous two-stage chemostat fermentations lasting over 1000 h, maintaining stable GFP titers, plasmid concentrations, and cell dry mass throughout the process. Our findings demonstrate that the auxotrophic marker thyA-based selection system, combined with strategic plasmid modifications, can substantially improve the genetic stability and productivity of E. coli in continuous bioprocesses. This approach provides a robust platform for sustainable, antibiotic- free production in industrial biotechnology, highlighting its potential for scale-up in long-term continuous fermentations.

|
Scooped by
mhryu@live.com
December 27, 11:05 AM
|
The dominant bacteria enriched in the Fusarium wilt plants’ rhizosphere are of increasing interest, as they adapt well to the diseased rhizosphere. However, general information about these bacteria is still lacking. Here, we perform a meta-analysis of Fusarium wilt plants rhizosphere and comprehensive studies to obtain information about the robust variation in the rhizosphere microbiome of Fusarium wilt plants. We demonstrate that Fusarium infection reproducibly changes the rhizosphere bacterial community composition. The rhizosphere microbiomes of Fusarium wilt plants are characterized by the enrichment of Flavobacterium, gene cassettes involved in antioxidant functions related to sulfur metabolism and the root secreted tocopherol acetate. We further isolate antagonistic Flavobacterium anhuiense from the diseased tomato rhizosphere, and reveal that the growth of F. anhuiense and the expression of genes related to carbohydrate metabolism in this strain are significantly stimulated by tocopherol acetate. Furthermore, the inhibitory effect of F. anhuiense against F. oxysporum and F. anhuiense population enhancement by tocopherol acetate are confirmed in planta. The robust variation in the rhizosphere microbiome elucidates key principles governing the general assembly mechanism of the microbiome in the Fusarium wilt plants’ rhizosphere. This study reveals how tomato plants recruit specific bacteria to combat Fusarium wilt. A key bacterium is identified to thrive on root-derived compounds and suppress the fungal pathogen, offering insights into plant-microbe defense alliances.

|
Scooped by
mhryu@live.com
December 27, 10:58 AM
|
Wolbachia is a ubiquitous endosymbiont in arthropods that produces small non-coding RNAs, which function as regulators in both the bacterium and its host. Although recent studies have shown cross-kingdom communication between Wolbachia and its host through Wolbachia-derived small non-coding RNAs (WsnRNAs), the functions of WsnRNAs have not been systematically examined. Here, we identify WsnRNAs in Wolbachia-infected Tetranychus truncatus Ehara via RNA-seq and investigate their impacts on host reproductive fitness. A total of 12 WsnRNAs were identified, along with their predicted precursors and hairpin structures. The predicted target genes of five highly expressed WsnRNAs are involved in reproductive development, as revealed by enrichment analysis. Inhibition of WsnRNA-744 and WsnRNA-3640 reduced fecundity, whereas inhibition of WsnRNA-6108 promoted it, indicating that different WsnRNAs exert opposing effects on host fecundity. These findings suggest that WsnRNAs mediate host-endosymbiont communication across species and could represent promising targets for Wolbachia-based pest control strategies.

|
Scooped by
mhryu@live.com
December 26, 3:48 PM
|
We describe peptide mapping through Split Antibiotic Resistance Complementation (SpARC-map), a method to identify the probable interface between two interacting proteins. Our method is based on in vivo affinity selection inside a bacterial host and uses high-throughput DNA sequencing to infer probable protein–protein interaction (PPI) interfaces. SpARC-map uses only routine microbiology techniques, with no reliance on specialized instrumentation, dedicated reagents, or reconstituting protein complexes in vitro. SpARC-map can be tuned to detect PPIs over a broad range of affinities, multiplexed to probe multiple PPIs in parallel, and its nonspecific background can be precisely measured, enabling the sensitive detection of weak PPIs. Using SpARC-map, we recover known PPI interfaces in the p21–PCNA, p53–MDM2, and MYC–MAX complexes. We also use SpARC-map to probe the purinosome, the weakly bound complex of six purine biosynthetic enzymes, where no PPI interfaces are known. There, we identify interfaces that satisfy structural requirements for substrate channeling, as well as protein surfaces that participate in multiple distinct interactions, which we validate using site-specific photocrosslinking in live human cells. Finally, we show that SpARC-map results can impose stringent constraints on machine learning–based structure prediction.

|
Scooped by
mhryu@live.com
December 26, 3:32 PM
|
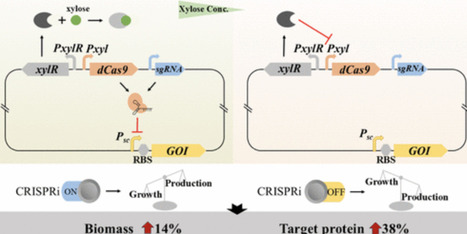
Bacillus subtilis is a critical host for protein production, with many industrial strains relying on strong constitutive promoters. However, this kind of promoter typically imposes a heavy burden on the host from the early stage of fermentation, leading to reduced growth rate and biomass. To overcome the drawbacks of these promoters, we developed a xylose-inducible CRISPRi module to dynamically control the activity of these promoters. The strength of this module was finely tuned via promoter engineering and the xylose concentration. The addition of xylose inhibited the target promoter and favored cell growth at an early stage, while the consumption of xylose recovered the strength of the promoter and facilitated protein expression, resulting in better balance between cell growth and protein production. The yield of a target protein was increased by 38% using this module. Our work provides a simple and effective method to upgrade industrial strains driven by strong constitutive promoters.
|

|
Scooped by
mhryu@live.com
Today, 12:42 PM
|
To address the growing emergence of multi-resistant phytopathogenic bacteria, innovative solutions are being explored in the field of plant health. Among them, bacteriocins, antimicrobial peptides or proteins secreted by bacteria, characterized by a highly specific spectrum of activity and involved in intra-specific competition, are gaining increasing interest. Bacteriocins can confer a positive selective advantage in both natural and agricultural environments, thereby contributing to microbiome modulation. Bacteriocin-producing rhizobacteria and lactic acid bacteria are already used as biocontrol agents against phytopathogenic bacteria, as well as plant growth stimulators. Bacteriocins can be produced in situ by using avirulent strains, or ex situ through industrial synthesis and applied as biopesticides. Nowadays, genetic engineering enables production of chimeric bacteriocins and their direct production in transgenic plants, avoiding the need for repeated treatments and limiting emergence of resistances. The selection of promising bacteriocins can be guided by omics-based approaches, notably metagenomics, which involve the direct extraction and sequencing of DNA from environmental samples and provides access to the genetic diversity in complex soil or plant-associated microbiomes. Combined with open-access databases and recently developed integrated tools, this approach not only facilitates the identification of known structures of bacteriocins, but also enables the prediction of potentially active peptides even those never experimentally characterized. Bacteriocin-based strategies, at the crossroads of molecular biology, microbial ecology and agronomy, hold significant potential for promoting sustainable agriculture through highly specific pathogen targeting. However, their large-scale implementation still faces several challenges, including standardization of strain screening protocols, compliance with regulatory frameworks and farmer acceptance.

|
Scooped by
mhryu@live.com
Today, 12:28 PM
|
Pure mycelium materials (PMMs) offer sustainable alternatives to traditional animal and synthetic polymer-based leather and petroleum-based packaging materials, by using innocuous fungi and agri-food biomass residues. Fungi respond readily to growth conditions and produce mycelia with tunable material properties to yield a diverse array of products. Additionally, these properties can be further modified through various post-processing techniques. This critical review examines how process engineering approaches, such as manipulating the fungal microenvironment, via optimal growth conditions, substrate selection, and bioreactor design, as well as post-processing techniques, can be effectively employed to engineer the properties of PMMs. It specifically describes the structure-property relationships of mycelium as governed by substrate assimilation and fermentation process parameters, and their impact on the fungal phenotype. To this, a comprehensive discussion on the role of fermentation conditions to mediate changes in fungal hyphal structure, composition, orientation, and cell wall composition – all of which determine the final mycelium material properties – is provided. By examining these relationships, this review aims to provide process insights that could guide the design and implementation of PMM production systems with enhanced yield and productivity along with the provision of tunable material properties. Finally, the review highlights other considerations such as life cycle assessment and regulatory requirements, and their relationships with process engineering, which are important for PMM development and their industry exploitation.

|
Scooped by
mhryu@live.com
December 27, 10:02 PM
|
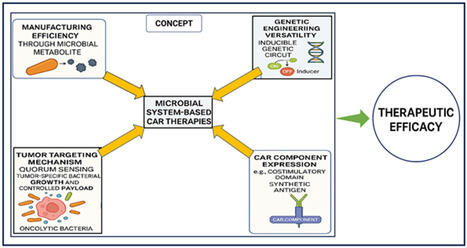
Chimeric antigen receptor (CAR)-based immunotherapies face significant translational challenges in solid tumor applications, particularly regarding manufacturing scalability, tumor targeting specificity, and antigen heterogeneity. This systematic review evaluates microbial systems as innovative platforms to address these limitations through synthetic biology-driven approaches, with a focus on bridging preclinical advances to clinical implementation. Analysis of 389 peer-reviewed studies (2015–2025) reveals that engineered probiotic strains (e.g., Escherichia coli Nissle 1917) achieve selective tumor colonization while functioning as programmable factories for: 1. Synthetic antigen production and single-chain variable fragment (scFv) expression, 2. Costimulatory domain delivery enabling antigen-agnostic CAR-T activation, 3. Tumor microenvironment modulation via immunostimulatory chemokines. Microbial platforms demonstrate superior manufacturing economics (70–90% cost reduction vs. conventional methods) and enhance CAR-T functionality through epigenetic reprogramming by microbial metabolites (e.g., short-chain fatty acids). CRISPR/Cas-engineered genetic circuits further enable precise spatiotemporal control of therapeutic payloads.Microbial systems represent transformative platforms for scalable, programmable CAR immunotherapy with significant potential for solid tumor targeting. Key barriers to clinical translation include biocontainment challenges, incomplete mechanistic understanding of tumor homing specificity, and safety validation requirements. Strategic integration of synthetic biology with microbial chassis offers a viable pathway toward accessible next-generation cancer therapies.

|
Scooped by
mhryu@live.com
December 27, 9:14 PM
|
PDBe-KB Complexes is a set of webpages that aggregate experimentally determined macromolecular complexes into unique complex-level records within the Protein Data Bank in Europe Knowledge Base framework. Each unique complex identifier unifies equivalent biological assemblies from the PDB that represent the same macromolecular complex, consolidating information on component identity, stoichiometry, symmetry, ligands, and PISA-derived assembly properties within a consistent framework. Relationships between complexes are established by comparing their compositions, capturing both sub-complexes that form part of larger ones with additional components, or higher-order complexes formed by repeating identical subunits. The current release of PDBe-KB Complexes includes over 100,000 unique complex compositions encompassing protein, nucleic acid, and mixed complexes. The SARS-CoV-2 spike protein complex serves as a representative case study, demonstrating how aggregation across hundreds of assemblies from different PDB entries reveals conformational diversity, ligand interactions, and antibody binding beyond traditional PDB entry-centric views. PDBe-KB Complexes provides a comprehensive foundation for comparative and functional analyses of macromolecular complexes across the PDB, supporting fundamental and translational research and education across life sciences. Users can explore PDBe-KB Complexes webpages; for example, the SARS-CoV-2 spike complex can be accessed via any corresponding PDB entries such as 9CXE at https://www.ebi.ac.uk/pdbe/pdbe-kb/complexes/9cxe.

|
Scooped by
mhryu@live.com
December 27, 12:36 PM
|
Ectomycorrhizal fungi form symbiotic relationships with a wide range of terrestrial plants, acquiring carbohydrates for themselves and promoting nutrient uptake in their host plants. However, some ectomycorrhizal fungi cannot effectively obtain the thiamine necessary for growth from their host or synthesize it themselves. Ectomycorrhizal fungi can recruit hypha-associated microorganisms, which play a vital role in promoting nutrient absorption and ectomycorrhizal root formation, ultimately colonizing within fruiting bodies to form a unique bacterial microbiota. In this study, non-targeted metabolomics and whole-genome sequencing were employed to investigate the colonization characteristics of the hyphae-associated bacterium Bacillus altitudinis B4 on the mycelial surface of ectomycorrhizal fungus Suillus clintonianus, as well as the synergistic promotion of thiamine synthesis and absorption by B. altitudinis B4 and the fungal mycelium, respectively. The results suggested that S. clintonianus first secreted ureidosuccinic acid and pregnenolone, recruiting the hyphae-associated bacterium B. altitudinis B4 to the mycelial surface. Subsequently, the ureidosuccinic acid secreted by S. clintonianus further stimulated B. altitudinis B4 to enhance thiamine production by increasing its biomass and upregulating the expression of related functional genes. Finally, S. clintonianus absorbed the thiamine secreted by the B. altitudinis B4, promoting fungal growth and increasing the colonization rate in association with Pinus massoniana. This study elucidates the thiamine acquisition mechanisms of ectomycorrhizal fungi, highlighting the critical role of bacterial partners in fungal nutrition and host-fungal interactions.

|
Scooped by
mhryu@live.com
December 27, 12:20 PM
|
Natural products (NPs) have long played a pivotal role in medicine, serving as both lead compounds and approved drugs in diverse therapeutic areas. Inspired by the 2004 review “The Role of Natural Product Chemistry in Drug Discovery”, we adopted a data-driven approach to assess the current role of NPs in drug discovery. We identified 119 NP-derived (NP-D) drugs, including 16 antibody-drug conjugates (ADCs), that were first approved globally between January 2000 and September 2025. Their development timelines, source organisms, manufacturing methods, indications, and routes of administration were analyzed. Six NP-D case studies illustrating successful progression from discovery to clinical use were highlighted: omaveloxolone (novel pharmacology, first-in-class), fidaxomicin, ibrexafungerp, epoxomicin, eribulin (known pharmacology, new drug class), and the gliflozin drug family (13 members). We also examined the presence and sales of NP-D agents among the top 200 brand name drugs in 2006, 2015, and 2024. Although overall NP-D drug sales have declined, dapagliflozin and empagliflozin demonstrate that blockbuster status is still achievable. While growth is modest, NPs remain highly valuable active pharmaceutical ingredients (APIs) with substantial clinical impact. Significant opportunities persist for drug-focused NP R&D efforts, as many recently validated therapeutic targets remain underexplored for NP-D chemotypes.

|
Scooped by
mhryu@live.com
December 27, 12:06 PM
|
l-Homoserine, a nonproteinic amino acid with extensive applications, holds significant potential for sustainable production via a cell factory, particularly with the nongrain feedstock glycerol. Here, Escherichia coli was systematically engineered to enhance l-homoserine production using glycerol as an alternative carbon source. Initially, we truncated homoserine kinase (ThrB) to avoid l-homoserine degradation and support cell growth. Subsequently, the main synthesis pathway was strengthened by limited-step gene overexpression and protein scaffold strategies. With further reprogramming of the glycerol assimilation pathway and heterologous introduction of the Entner–Doudoroff pathway to balance cofactor supply, an l-homoserine hyperproducer HZ23/p99ArhtB could achieve a titer of 90.21 g/L, with a yield of 0.49 g/g glycerol and a productivity of 1.50 g/L/h. Our study demonstrated the potential of glycerol as a sustainable feedstock for high-yield production of l-homoserine and provides a robust framework for the industrial-scale production of this important precursor.

|
Scooped by
mhryu@live.com
December 27, 11:49 AM
|
Conventional genetic engineering approaches for bacterial metabolic pathway manipulation, although highly applicable, still face limitations including metabolic burden, irreversibility, and dependency on host cellular machinery. Cell-penetrating peptide-peptide nucleic acid conjugates (CPP-PNAs), known for their applicability as antibacterial tools and in the elucidation of protein function, offer a promising alternative to overcome such limitations. Since the application of CPP-PNA in metabolic engineering and pathway elucidation remains largely unexplored, we developed and validated a CPP-PNA platform using Synechocystis sp. PCC 6803 as a model system to demonstrate targeted metabolic pathway evaluation. High compatibility and dose-dependent permeation efficiency in strain PCC 6803 was first observed when the amphipathic CPP (KFF)₃K was employed, achieving clear cell growth inhibition at 10 µM and above. Specific targeting of D-lactate dehydrogenase (Ddh) using CPP-Syn6803ddh conjugates achieved near-complete protein translation knockdown within 24 h, as confirmed by Western blot analysis. Metabolomics analysis using LC-MS on predetermined metabolites revealed that CPP–PNA treatment produced metabolic effects comparable to stable genetic knockout strains, with both approaches showing a significant 2.5-fold increase in pyruvate accumulation compared to wild-type controls. Further elucidating the reason for pyruvate accumulation, we observed compensatory activation of the glyoxalase pathway at 48 h post-treatment, resulting in 3-fold increased D-lactate production presumably through methylglyoxal detoxification. Validating this observation, RT-qPCR analysis confirmed 2-3-fold upregulation of the glyII gene, encoding for the glyoxalase II (GlyII) enzyme, in both CPP-PNA treated and knockout strains, while double CPP-PNA inhibition experiments targeting both Ddh and glyoxalase pathways suppressed D-lactate accumulation. This study establishes CPP-PNAs as efficient tools for rapid, and simple metabolic pathway investigation. The approach produces results comparable to conventional genetic knockouts while offering dose-dependent control and avoiding permanent genomic alterations. Our findings reveal unexpected metabolic complexity in Synechocystis sp. PCC 6803 D-lactate synthesis under light conditions and demonstrate the utility of CPP-PNA for uncovering compensatory pathway activation. This platform represents a valuable addition to bacterial genetic engineering, addressing some of the critical limitations faced by conventional approaches, while showing potential for further understanding the biochemistry of metabolite-producing bacteria.

|
Scooped by
mhryu@live.com
December 27, 11:26 AM
|
Land plant evolution has been marked by bursts of novelty, often underpinned by extensive genomic innovation. A key mechanism driving these changes is horizontal gene transfer (HGT), the process by which genes move between species and even across taxonomic kingdoms. HGT can accelerate evolutionary change through the rapid introduction of new genes yet its importance in plant biology is only beginning to be understood. Here, we review the functional contributions of HGT during the origin and diversification of land plants. We discuss the occurrence of HGT throughout plant evolution and its impact on the origin of defining traits from cell walls to developmental programs. Beyond ancient contributions, HGT continues to drive the emergence of lineage-specific innovations. Recently acquired bacterial and fungal genes make complex functional contributions to processes including stress response, pathogen defence, and development across plant phylogeny. These observations suggest that HGT was, and continues to be, a major force shaping plant evolution, exemplifying the potential significance of HGT in eukaryotic biology more broadly.

|
Scooped by
mhryu@live.com
December 27, 11:03 AM
|
The use of nucleic acid-based nanostructures as synthetic biological tools to interface with and regulate cell processes remains challenging. A major obstacle lies in nuclear delivery and retention within live eukaryotic cells. Here, we present a platform of single-stranded RNAs that can co-transcriptionally fold into defined nanostructures and assemble into rings, ribbons, and nanonet-like architectures. We validate the formation of these structures in vitro using atomic force microscopy. Then, we demonstrate the functional integration of fluorescent aptamers and RNA sensing capability within the single chain by co-folding with these structures. Notably, we show that the RNA nanonets can be co-transcriptionally produced and assembled directly inside the nucleus of live human cells. We use confocal live-cell imaging and transmission electron microscopy to reveal well-defined nanostructure patterns retained in the nucleus. Together, these results establish a genetically encoded, self-assembling RNA nanostructure system with programmable geometry and localization, providing a foundation for the development of RNA-based nanodevices to examine biological properties in live cells and tissues. The use of nucleic acid-based nanostructures as synthetic biological tools remains challenging. Here they present synthetic RNA structures that fold and self-assemble inside human cell nuclei, offering a platform for imaging, sensing, and future therapeutic applications.

|
Scooped by
mhryu@live.com
December 26, 3:57 PM
|
Single-cell sequencing methods uncover natural and induced variation between cells. Many functional genomic methods, however, require multiple steps that cannot yet be scaled to high throughput, including assays on living cells. Here we develop capsules with amphiphilic gel envelopes (CAGEs), which selectively retain cells and large analytes while being freely accessible to media, enzymes and reagents. Capsules enable high-throughput multistep assays combining live-cell culture with genome-wide readouts. We establish methods for barcoding CAGE DNA libraries, and apply them to measure persistence of gene expression programs in cells by capturing the transcriptomes of tens of thousands of expanding clones in CAGEs. The compatibility of CAGEs with diverse enzymatic reactions will facilitate the expansion of the current repertoire of single-cell, high-throughput measurements and their extension to live-cell assays.

|
Scooped by
mhryu@live.com
December 26, 3:36 PM
|
Precision fermentation uses microorganisms (e.g., yeast, bacteria, or fungi) to produce ingredients such as food proteins. These proteins are promising animal-free alternatives due to their ability to better mimic the texture and taste of animal-derived products than do plant proteins. However, conventional purification methods, such as chromatography, are costly and designed for high-purity, high-value products. Cost-effective production of bulk food proteins (e.g., milk or egg proteins) requires alternative downstream approaches. This review explores more affordable processing strategies suitable for recombinant food proteins. Emphasis is placed on achieving ingredient functionality, such as emulsifying, foaming, and gelation, over purity to reduce energy use and material losses. Alternative methods, including coacervation with food-grade polyanions, are discussed. Some approaches focus on the unique properties of food proteins, such as the calcium sensitivity of α- or β-caseins, to enable simplified extraction. Many of these strategies are at the conceptual stage and require further research.
|






 Your new post is loading...
Your new post is loading...

























elm, interactions between mycelium cell wall components - proteins, glucan, and chitin - and additives such as plasticizers (glycerol, sorbitol) and cross-linkers (citric acid, tannic acid)