Your new post is loading...

|
Scooped by
?
Today, 2:00 AM
|
Rhizosphere microorganisms are known to be able to modify the plant's ability to resist abiotic stresses. It is, however, difficult to modify microbial communities to improve plant phenotypes. Here we tested if a rhizosphere microbial community from a water-stress naive soil could be modified by adding DNA extracted from soils with a water stress history. Six-week-old wheat plants growing under low or high-water availabilities were inoculated with DNA extracted from soils with contrasting long-term histories of water availability - one continuously and the other intermittently exposed to water deficit. The fate of the inoculated DNA in the rhizosphere microbial communities was assessed by shotgun metagenomics. Putatively transferred inoculum genes were disproportionately found in the Acidobacteria and Bacteroidetes and belonged to functional category such as antibiotics, biofilm, and carboxylates metabolism, among others. These functional categories were shared by pre-inoculation laterally transferred genes in the recipient soil, highlighting their usefulness for life in soil. The "continuous" inoculum reduced the stress levels of wheat under reduced soil water content, suggesting that the natural genetic transformation of the rhizosphere community can feedback to the plant. Altogether, we are providing evidence for an ecological mechanism that could be harnessed to modify plant-associated microbial communities and help plants sustain water stress.

|
Scooped by
?
Today, 1:56 AM
|
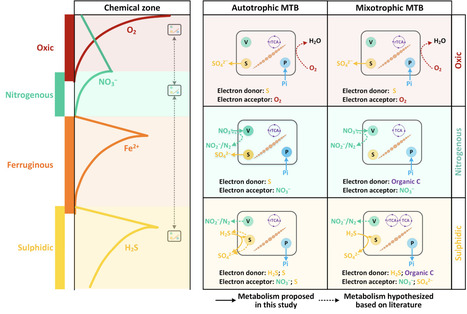
Magnetotactic bacteria (MTB) are capable of accumulating and storing intercellular pools containing phosphorus, sulfur, nitrogen and carbon. Yet, the metabolic pathways connected with their intracellular storage of polyphosphate, elemental sulfur, nitrate, and calcium carbonate, and the environmental influence on MTB inclusions and their quantitative contribution to sedimentary chemical budgets remain underexplored. Using a combination of chemical characterization, ultrastructural and compositional analyses, and phylogenetic and genomic insights into microbial assemblages, we investigated the influence of geochemical parameters on the diversity, ecophysiology, and distribution of MTB in freshwater and brackish sediments. Special focus is given to intracellular inclusions and their metabolic pathways to uncover functional traits and ecological roles in elemental cycling. Of the 118 examined MTB cells, polyphosphate granules (75%) and nitrate-containing vacuoles (67%) were the most common inclusions followed by sulfur globules (25%) and calcium carbonate granules (8%). The intracellular phosphorus, nitrogen, and sulfur stored in MTB cells were conservatively estimated to account for 0.60%, 0.91%, and 0.23% of the total sedimentary P, N, and S in the investigated freshwater sediments, respectively. The ubiquitous nature and important ecological role of MTB can be explained by their ability to sequester chemical elements into intracellular inclusions, giving them a metabolic advantage in dynamic chemically stratified environments. The extraordinary phylogenetic diversity of MTB, coupled with their capability to hyperaccumulate and store a wide range of elements and compounds, represents a significant resource for biotechnology innovation.

|
Scooped by
?
Today, 1:45 AM
|
The Gene Ontology (GO) knowledgebase https://geneontology.org is a comprehensive resource describing the functions of genes. The GO knowledgebase is regularly updated and improved. We describe here the major updates that have been made in the past 3 years. The ontology and annotations have been expanded and revised, particularly in several areas of biology: cellular metabolism, multi-organism interactions (e.g. host-pathogen), extracellular matrix proteins, chromatin remodeling (e.g. the “histone code”), and noncoding RNA functions. We have released version 2 of a comprehensive set of integrated, reviewed annotations for human genes, which we call the “functionome.” We have also dramatically increased the number of GO-CAM models, with over 1500 models of metabolic and signaling pathways, primarily in human, mouse, budding and fission yeast, and fruit fly. Finally, we discuss our current recommendations and future prospects of AI in the use and development of GO.

|
Scooped by
?
Today, 1:41 AM
|
Colouration of textiles is responsible for 3% of global greenhouse gas emissions and 20% of global wastewater generation. An estimated 20% of the dyes and pigments used in textile colouration is lost to wastewater. Synthetic dyes and pigments, along with many of the chemicals added to their formulations, are nonbiodegradable, persistent, and bioaccumulative in aquatic ecosystems, posing signi cant risks throughout the food chain. Regulatory bodies in both Europe and the United States are continuing to ban, restrict, and phase out many synthetic dyes and pigments. The EU recently banned a significant number of common textile dyes known to release carcinogenic compounds, while in the United States, 8 common synthetic food dyes are being targeted for immediate ban in the US food supply, with the FDA planning to phase out all remaining synthetic food dyes. While bio-based alternatives are available on the market, they are currently more expensive, restricting their application to niche product applications that can absorb the price premium. The total market size for dyes and pigments is more than US$ 50 billion and is growing at a rate of 3–10% annually. The ‘cost of being unsustainable’ is increasing rapidly, particularly in the EU, as new regulations make the use of hazardous chemicals more difficult and expensive. This is driving a shift toward the use of more sustainable chemicals.

|
Scooped by
?
Today, 1:31 AM
|
Recent efforts in bottom-up synthetic biology focus on fabricating programmable biological units that can be viewed as synthetic cells. Combining microfluidic techniques with cell-free protein expression systems defines the geometrical limits of the synthetic cell (e.g. microfluidic compartments, droplets, vesicles) and facilitates communication pathways to distribute functions over an assembly of synthetic cells. In this review, we describe and compare the different strategies implemented to reconstitute cell–cell communication among synthetic cells. We focus especially on various experimental setups of microcompartmentalization containing a cell-free expression system and genetic material. We highlight efforts to develop and engineer different modes of communication among the synthetic cells in different forms, varying by the degree of permeability, resource renewal, stability, and scalability, and how these influence the trade-off between programmability and biomimicry. We then summarize recent progress in the realization of different stages of communication (signaling, processing, and output generation) by genetic circuits, holding great promise for applications in synthetic biology and biotechnology.

|
Scooped by
?
Today, 1:27 AM
|
Single-stranded RNA (ssRNA) can activate the mechanosensitive ion channel Piezo1 in the gut. However, its source and role in colorectal cancer (CRC) remain unclear. In the present study, we demonstrate that ssRNA within fecal extracellular vesicles (FEVs) derived from bacteria, particularly those susceptible to lysis by ursodeoxycholic acid (UDCA), can suppress CRC progression via Piezo1 activation. Gut-specific Piezo1-knockout mice developed more tumors following CRC induction. Similarly, Piezo1-deficient CRC cell lines exhibited increased proliferation with upregulated Wnt/β-catenin signaling. Mechanistically, Piezo1 activation downregulated the transcription factor Zfp281, decreasing the expression of its target gene Lgr5 and dampening Wnt/β-catenin signaling. Notably, oral UDCA administration enhanced FEV rupture, increasing luminal “naked” ssRNA and mitigating high-fat diet-induced CRC in vivo. These findings identify the bacterial ssRNA-Piezo1 axis as a potential therapeutic target in CRC.

|
Scooped by
?
Today, 1:19 AM
|
Under severe nutrient-limiting conditions, Bacillus subtilis is able to form highly resilient endospores for survival. However, to avoid this irreversible process, it employs an adaptive strategy termed cannibalism, a form of programmed cell death, to outcompete siblings and delay sporulation. One of the three cannibalism toxins, the epipeptide EPE, is encoded by the epeXEPAB operon. The pre-pro-peptide EpeX undergoes post-translational modification and processing to be secreted as the mature EPE toxin. While EPE production is tightly regulated at multiple levels, this study focuses on the post-transcriptional control by the small regulatory RNA FsrA, which is transcriptionally regulated by the global iron response regulator Fur. Electrophoretic mobility shift assays and RNA structure probing revealed two binding sites of FsrA within the intergenic region between epeX and epeE flanking the annotated epeX terminator structure and potentially interfering with RNA stability and epeXEP expression. Reporter assays revealed decreased levels of EPE-dependent stress response in the absence of FsrA, indicative of a positive FsrA effect on gene expression under iron-limited conditions; in contrast to the normally inhibitory activity of FsrA. Together, our findings suggest that under iron starvation, FsrA promotes RNA processing and enables epeE translation, ultimately enhancing EPE production.

|
Scooped by
?
Today, 12:30 AM
|
Base editors enable precise genome modification but are constrained by bystander edits that limit their applicability. Existing strategies to enhance precision often compromise efficiency and remain highly sequence dependent. Here we present a parallel engineering approach that optimizes both guide RNAs and the deaminase enzyme to minimize bystander editing without sacrificing activity. We designed a library of 3′-extended guide RNAs and identified context-dependent variants that improved specificity. Using a precision-driven phage-assisted evolution system and protein language models, we evolved adenine base editor variants two- to threefold more precise than adenine base editor ABE8e while maintaining high efficiency across a library of thousands of human pathogenic contexts in vitro. Our findings establish a scalable framework for precision engineering of base editors, addressing a major challenge in genome editing. Base editors are made more precise through deaminase and gRNA engineering.

|
Scooped by
?
Today, 12:12 AM
|
Riboswitches are structured allosteric RNA molecules that change conformation upon metabolite binding, triggering a regulatory response. Here we focus on the de novo design of riboswitch-like aptamers, the core part of the riboswitch undergoing structural changes. We use Restricted Boltzmann machines (RBM) to learn generative models from homologous sequence data. We first verify, on four different riboswitch families, that RBM-generated sequences correctly capture the conservation, covariation and diversity of natural aptamers. The RBM model is then used to design new SAM-I riboswitch aptamers. To experimentally validate the properties of the structural switch in designed molecules, we resort to chemical probing (SHAPE and DMS), and develop a tailored analysis pipeline adequate for high-throughput tests of diverse sequences. We probe a total of 476 RBM-designed and 201 natural sequences. Designed molecules with high RBM scores, with 20% to 40% divergence from any natural sequence, display ≈ 30% success rate of responding to SAM with a structural switch similar to their natural counterparts. We show how the capability of the designed molecules to switch conformation is connected to fine energetic features of their structural components. Riboswitches are allosteric RNA molecules that change conformation upon ligand binding to regulate downstream genes. Here, the authors use Restricted Boltzmann machines trained on natural sequences to design new riboswitch aptamer domains, and validate their functionality via chemical probing.

|
Scooped by
?
December 18, 11:48 PM
|
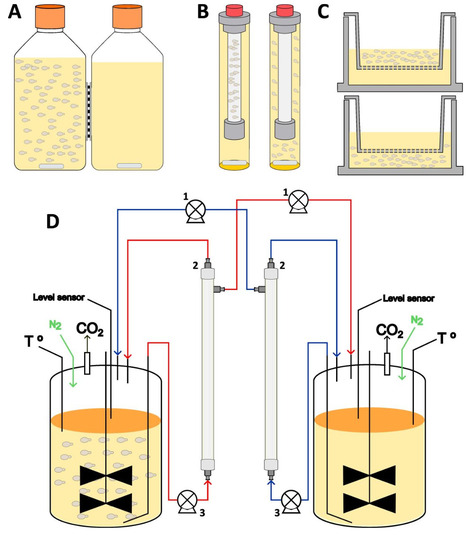
With some exceptions, wine fermentation has increasingly relied on the use of Saccharomyces cerevisiae starters since the last decades of the past century. However, there is growing interest on the understanding of spontaneous wine fermentation, as well as on the use of complementary non-Saccharomyces wine starters. This in turn raises the question of the importance of interspecific interactions in winemaking and the underlying mechanisms. An important question about these interspecies recognition mechanisms is whether or not it is mediated by physical contact between yeast cells. To address this topic, different laboratories have developed diverse devices to cultivate at least two yeast species in the same growth medium without cell-to-cell contact between them. In this work, we compared four of the most popular systems and found that one of them (twin-bottles exchange through a flat membrane) showed very limited metabolite exchange. Among the other systems (Transwell, dialysis tube, or active exchange through hollow fiber cross-flow filtration devices), each one showed specific characteristics that made them more or less suitable, depending on the objectives of each experiment. The option showing the best versatility and efficiency was the use of active exchange. Our results highlight the importance of carefully characterizing the compartmentalization system when drawing conclusions about the impact of cell-to-cell contact in fermentation experiments.

|
Scooped by
?
December 18, 11:30 PM
|
Anaerobic ammonium oxidizing bacteria in the class “Candidatus Brocadiia” in the Planctomycetota are the only known group of bacteria capable of producing energy by coupling the oxidation of ammonium to the reduction of nitrite within a unique bacterial organelle called the anammoxosome. Due to the lack of homologs in other species, it is hypothesized that the key enzyme in this process, the hydrazine synthase complex, originated by de novo birth. We performed extensive searches for proteins that exhibited similarity in sequence and structure to the hydrazine synthase subunits and identified distantly related homologs in anaerobic bacteria from the phyla Planctomycetota and Desulfobacterota. However, key residues of importance for the enzymatic function were not conserved, rejecting the hypothesis that the identified genes represent previously unrecognized anammox bacteria. Phylogenetic analyses indicate that the anammox pathway has been assembled from genes acquired by horizontal gene transfer from a variety of anaerobic bacteria. The ancestral states of enzymes in the hydroxylamine oxidoreductase family were inferred, and transitions between reductive and oxidative forms of the enzymes were mapped onto the phylogenetic tree. Finally, it is shown that the signal sequences of key enzymes in the anammox pathway are able to transport a reporter gene into the periplasm of Escherichia coli cells. In conclusion, our findings suggest that the hydrazine synthase complex has evolved from already existing heme-binding periplasmic proteins and that the anammoxosome has an endogenous origin.

|
Scooped by
?
December 18, 9:51 PM
|
The impressive capabilities of natural pattern recognition systems have inspired their synthetic recreation for many chemical and biological applications. However, developing artificial receptors for pattern recognition is currently constrained by a laborious trial-and-error process within a limited selection space of synthetically generated molecules/materials. Here, we propose pattern recognition aptamers (PRAs)─a set of single-stranded nucleic acid ligands with quasi-specificity for multiple targets─that can be evolved through systematic exponential enrichment from a nucleic acid library for high-precision target identification. Our approach allows for the reliable generation of customized artificial receptors over a limited number of selection rounds. Using bacteria as model analytes, we developed 9 PRAs targeting 15 common bacteria through 3 rounds of evolutionary screening, achieving an identification accuracy of 98.5% in blinded unknown bacterial identification. This approach provides a generalized pipeline for creating customized pattern recognition arrays, supporting their potential to meet rapidly increasing application demands.

|
Scooped by
?
December 18, 6:31 PM
|
The convergence of synthetic biology and biosensor technology is driving a scientific and technological revolution in biotechnology. Characterized by its interdisciplinary nature, synthetic biology applies engineering principles to design and construct novel biological components and systems, significantly advancing biosensor development. As a key interface between the digital and biomolecular worlds within synthetic biology, cell-free biosensors (CFBs) leverage core advantages─including rapid prototyping, high controllability, excellent biosafety, and tolerance to potentially toxic substances─to demonstrate increasingly significant strategic value and broad application potential in numerous frontier fields. These fields encompass the precise design of biological circuits, intelligent optimization of metabolic pathways, and efficient in vitro diagnostics. CFBs overcomes limitations inherent in traditional cell-based sensors, offering a powerful platform for instant, on-demand biomolecular detection. This review aims to explore the innovative potential unlocked by the integration of synthetic biology and biosensors and to systematically summarize the latest research progress and technological breakthroughs in the CFB field.
|

|
Scooped by
?
Today, 1:58 AM
|
Type IV secretion systems (T4SS) enable the spread of antibiotic resistance and other virulence factors. In Gram-positive bacteria, T4SSs have long been thought to lack VirB2-like proteins that form conjugative pili and instead rely on adhesins for cell-cell contacts. Yet, it has remained unclear how subsequent DNA transfer (conjugation) is physically mediated. Here we identify a VirB2-like protein, PrgFB2, from the clinically isolated conjugative plasmid pCF10 in Enteroccocus faecalis and show that it is essential for conjugation. Structural modeling confidently predicts a pilus-like assembly for PrgFB2. We validate this prediction through mutagenesis, conjugation assays, and targeted chemical labeling. By combining various machine learning bioinformatic techniques, we analysed >1000 Gram-positive conjugative plasmids from diverse species, including major pathogens, and identified pili forming VirB2-like proteins in almost all of them. Our findings overturn the prevailing view that Gram-positive T4SSs lack pili. This discovery provides a new framework for understanding horizontal gene transfer and highlights critical targets for combating antimicrobial resistance and virulence in Gram-positive bacteria.

|
Scooped by
?
Today, 1:51 AM
|
Prime editing (PE) enables the precise installation of intended base substitutions, small deletions or small insertions into the genome of living cells. While the use of Cas9 nickase can avoid DNA double-strand breaks (DSB), undesired insertions and deletions (indels) often accompany the correct edits, particularly when PE activity increased. Here we show that the anti-CRISPR (Acr) protein AcrIIA5 can significantly enhance PE activity by up to 8.2-fold while markedly reducing byproduct indels. Further investigation reveals that AcrIIA5 can promote PE across various approaches (PE2, PE3, PE4, PE5, and PE6), edit types (substitutions, insertions and deletions), and endogenous loci. Mechanistically, AcrIIA5 appears to inhibit the re-nicking activity of PE complex rather than enhancing the core editing machinery itself, suggesting a distinct mode of interaction with Cas9. Overall, we demonstrate that a known “inhibitor” Acr protein can unexpectedly acting as an “enhancer” of CRISPR/Cas-based genome editing, providing an effective strategy to optimize PE specificity and activity. Prime editing enables precise genetic modification but often suffers from unwanted byproducts. Here, authors show that the anti-CRISPR protein AcrIIA5 unexpectedly enhances prime editing efficiency while reducing unintended indels, offering an effective strategy to improve activity and specificity.

|
Scooped by
?
Today, 1:43 AM
|
Directed evolution facilitates functional adaptations through stepwise changes in sequence that alter protein structure. While most campaigns yield solutions that maintain the framework of a rigid protein architecture, a few have produced enzymes with more notable structural differences. One example is a polymerase that was evolved to synthesize threose nucleic acid (TNA) with near-natural activity. Understanding how this enzyme arose provides a model for studying pathways that guide enzymes toward more productive regions of the fitness landscape. Here, we trace the evolutionary trajectory of an unnatural polymerase by solving crystal structures of key intermediates along the pathway and evaluating their biochemical activity. Contrary to the view that fidelity is a product of increased catalytic efficiency, we find that accuracy and catalysis are decoupled activities guided by separate ground-state and transition-state discrimination events. Together, these results offer a glimpse into the forces responsible for shaping the emergence of new enzyme functions. Engineering polymerases to synthesize alternative genetic polymers remains a challenging problem in synthetic biology. The current study offers insights into the structural and biochemical changes responsible for improving the fidelity and catalytic activity of a laboratory evolved TNA polymerase.

|
Scooped by
?
Today, 1:35 AM
|
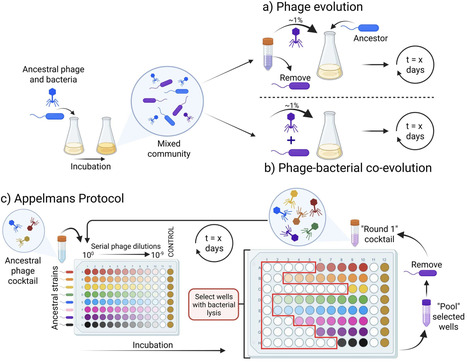
Bacteriophages are viruses that selectively prey on bacteria. Their use in treating antimicrobial-resistant bacterial infections is steadily increasing due to the need for alternative therapies. The application of phage therapy is not without its challenges, including difficulties associated with isolating phages against a target strain, the limited infectivity of a phage, the cost and complexity of producing well-characterized phage stocks, and the emergence of phage resistance. The directed adaptation of phage to a specific bacterial target, also known as ‘phage training’, leverages the natural evolutionary capacity of phages and can be used to bolster their bacterial killing abilities. Phage training dates back almost as far as phage therapy itself, being used to expand the therapeutic use of phages. Numerous reports showcase the success and benefits of phage training in vitro and its potential to operate effectively within the framework of phage therapy. However, the time needed to train a given phage, followed by genotypic and phenotypic characterisation of both pre- and post-trained phages, is a major limitation. Here, we explore oversights of the phage training process and propose some considerations and solutions to help drive the field forward to enable its feasible integration into phage therapy.

|
Scooped by
?
Today, 1:30 AM
|
Persister cells survive any severe stress including antibiotics, starvation, heat, oxidative conditions, and phage attack, by entering a dormant physiological state. They arise without genetic change and can resume growth once the stress is removed and nutrients are available. Critically, upon resuscitation, persister cells can reconstitute infections. Although it is known that persister cells resuscitate in proportion to their ribosome content, it has remained unclear whether ribosome levels also influence the formation of persister cells. Here, we used fluorescence-activated cell sorting (FACS) to fractionate exponentially growing cells into four populations spanning low to high ribosome levels and demonstrated that cells with low ribosome content form persister cells approximately 80-fold more frequently than cells with population-average ribosome levels. These findings show that persister cell formation is inversely proportional to ribosome abundance. Cells with low ribosome levels are less metabolically-active and therefore less capable of initiating a stress response like most cells; instead, they become dormant.

|
Scooped by
?
Today, 1:21 AM
|
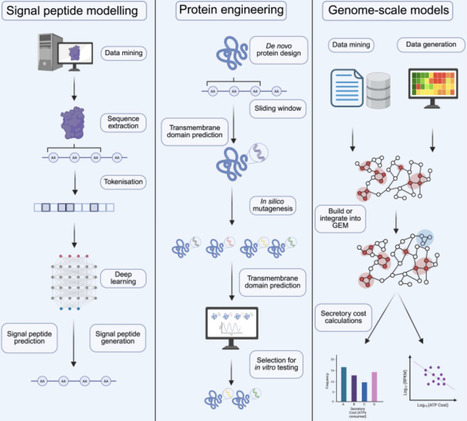
Protein secretion in mammalian cells is the active transport of proteins from the cytoplasm to the extracellular space. It plays a fundamental role in mammalian physiology and signaling, as well as biotherapeutics production and cell and gene therapies. The efficacy of protein secretion, however, is impacted by features of the secreted protein itself, and the host-cell machinery that supports each step of the secretion process. High-throughput techniques such as microfluidics, cell display, and cell encapsulation assays for the study and engineering of secreted proteins are transforming biomedical knowledge and our ability to modulate protein secretion. In addition, computational advances, including signal peptide modeling, whole-protein machine learning models, and genome-scale simulations, are opening new pathways for rational design of protein secretion. Here, we highlight recent developments in secretion engineering that are leading to the convergence of high-throughput experimentation and machine learning methods and can help address current challenges in bioproduction and support future efforts in cell and gene therapy while enabling new modalities.

|
Scooped by
?
Today, 1:15 AM
|
Extracting fungal hyphae with their naturally associated microbiota from soil samples presents a significant challenge due to their small size, typically in the micrometer range, and the formation of dynamic fungal networks. We combined elements of previous protocols and automated the wet-sieving steps of the methodology to efficiently extract fungal hyphae from various soil types, including natural loamy soils. This approach reduces manual handling, minimizes operator-dependent variability, and shortens processing time by up to 2.5-fold. Unlike earlier methods that require sand or glass bead supplementation, which can introduce artificial conditions and limit large-scale field applications, our Sieving and Sucrose Centrifugation (SSC) method avoids these drawbacks. The SSC technique enables both quantification of hyphal length density (HLD) and, importantly, preserves surface-associated microbes for downstream analyses. Among the tested methods, SSC yielded the highest hyphal length density. Using a combination of microscopy, molecular techniques, and next-generation sequencing (NGS), we demonstrate that this method allows targeted study of bacteria tightly attached to fungal hyphae. Furthermore, the SSC approach effectively enriched fungal hyphae from a highly diverse soil community, establishing a dependable tool for advancing research on fungal hyphae as microbial hotspots in soil ecosystems.

|
Scooped by
?
Today, 12:22 AM
|
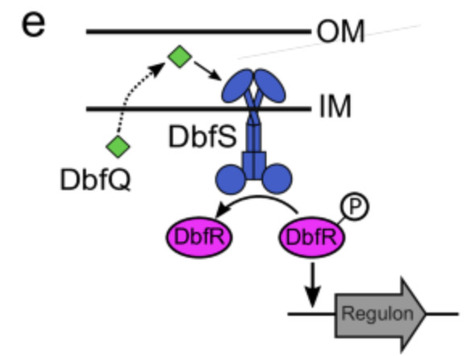
Two-component signaling pathways allow bacteria to sense and respond to environmental changes, yet the sensory mechanisms of many remain poorly understood. In the pathogen Vibrio cholerae, the DbfRS TCS controls the biofilm lifecycle, a critical process for environmental persistence and host colonization. Here, we identified DbfQ, a small periplasmic protein encoded adjacent to dbfRS, as a direct modulator of pathway activity. DbfQ directly binds the sensory domain of the histidine kinase DbfS, shifting it toward phosphatase activity and promoting biofilm dispersal. In contrast, outer membrane perturbations, caused by mutations in lipopolysaccharide biosynthesis genes or membrane-damaging antimicrobials, activate phosphorylation of the response regulator DbfR. Transcriptomic analyses reveal that DbfR phosphorylation leads to broad transcriptional changes spanning genes involved in biofilm formation, central metabolism, and cellular stress responses. Constitutive DbfR phosphorylation imposes severe fitness costs in an infection model, highlighting this pathway as a potential target for anti-infective therapeutics. We find that dbfQRS-like genetic modules are widely present across bacterial phyla, underscoring their broad relevance in bacterial physiology. Collectively, these findings establish DbfQ as a new class of periplasmic regulator that influences TCS and bacterial adaptation. A TCS, DbfRS, regulates biofilm formation in Vibrio cholerae. Here, Nguyen et al. identify a small periplasmic protein that controls the activity of the system’s receptor, and show that DbfRS responds to membrane stress and regulates additional processes such as metabolism and cell envelope biosynthesis.

|
Scooped by
?
Today, 12:03 AM
|
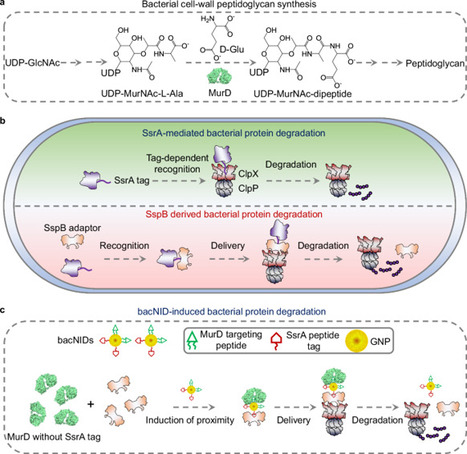
Antibiotic resistance stands as a formidable global challenge to public health. Herein, we present a bacterial nanoinducer (bacNID) designed for targeted protein degradation in treating bacterial infections. Specifically, bacNID is engineered by grafting targeting peptides of MurD and SspB onto gold nanoparticles (GNPs). MurD plays a pivotal role in peptidoglycan production for cell wall synthesis, while SspB recruits SsrA-tagged proteins for degradation by ClpXP protease. The effectiveness of bacNIDs in targeted MurD degradation via ClpXP is demonstrated across both Gram-positive and Gram-negative bacterial strains. Importantly, prolonged exposure to bacNIDs does not result in the acquisition of resistance in either Staphylococcus aureus (S. aureus) or Salmonella typhimurium (S. typhimurium), even after 25 successive treatment passages. This stands in stark contrast to the rapid emergence of robust resistance observed with norfloxacin, evidenced by a 243-fold reduction in antibacterial activity against S. aureus after just 15 passages, and a 7410-fold decrease in activity against S. typhimurium over 22 passages. Moreover, the antimicrobial potential of bacNIDs is evaluated in vivo using S. aureus-infected nonhealing skin and corneal wounds. In summary, this study unveils a potent nanotechnology-driven strategy for targeted bacterial protein degradation with promising implications for in vivo antimicrobial applications. In this work, authors present a nanoinducer (bacNID), which enables targeted bacterial protein degradation via ClpXP, leading to the effective treatment of bacterial infection (Staphylococcus aureus and Salmonella typhimurium) without inducing resistance.

|
Scooped by
?
December 18, 11:35 PM
|
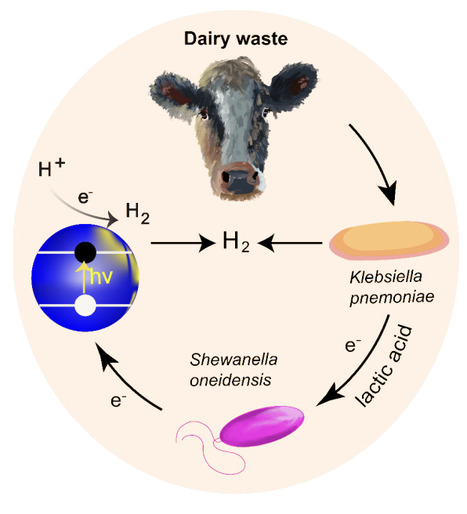
Dairy waste, such as whey resulting from cheese production, is produced in massive volumes worldwide and is regarded as environmentally difficult to dispose of due to its high organic content. Harnessing the potential of this waste material to support the synthesis of valuable products such as hydrogen fuel or reduced graphene oxide, which may be utilized for conductive thin films and energy storage can reduce waste and add revenue streams for dairy farmers. Here, we demonstrate a circular bioeconomy using alginate-encapsulated co-cultures of Shewanella oneidensis together with lactic-acid-producing bacteria Klebsiella pneumoniae. These co-cultures can directly metabolize unprocessed cheese-making waste as an electron source instead of costly, environmentally high-impact lactic acid. Alginate-encapsulated co-cultures fed unprocessed dairy waste showed a 2-to-3-fold higher graphene oxide reduction rate compared to S. oneidensis monocultures with no supplemental electron source. Encapsulated co-cultures were able to be recycled for more than 30 days with no measurable decrease in graphene oxide reduction efficiency, showing compatibility with future industrial scaling. Photocatalytic hydrogen generation with cadmium selenide quantum dots as the catalyst resulted in 6-fold increases in hydrogen produced by co-cultures using milk as an electron source precursor for the system in comparison to S. oneidensis monocultures without any added electron sources. Thus, dairy waste may be processed to drive the synthesis of valuable products utilizing microbial electron transfer processes, converting a significant fluvial environmental pollutant into a valuable renewable energy resource that could provide a robust alternative revenue stream for dairy farmers in a volatile industry.

|
Scooped by
?
December 18, 10:04 PM
|
The production of lactic acid, a crucial platform chemical by microbial fermentation, is currently hindered by its low production efficiency. Herein, this study aims to enhance the synthetic efficiency of lactic acid in widely used Komagataella phaffii by reconfiguring the synthetic pathway and a metabolite damage-repair system. First, through the introduction of lactic acid dehydrogenase (BsLDH) from Bacillus subtilis and the transporter LutP from Bacillus coagulans, the titer of lactic acid was increased from 1.5 g/L in the original strain to 55.4 g/L using glucose. In particular, the titer was improved to 87.2 g/L by introducing metabolite damage-repair genes for NAD(P)HX detoxification and phosphate-based inhibitor elimination. In addition, the carbon source transport and metabolism pathway were strengthened, resulting in titers of 18.7 and 7.7 g/L from glycerol and methanol. Finally, the strains were scaled up in a 5 L bioreactor, achieving lactic acid titer values of 153.0, 133.2, and 37.4 g/L from glucose, glycerol, and methanol, respectively. This study significantly improved the yield of lactic acid production from low-cost carbon sources by microbial fermentation, demonstrating the potential of engineered K. phaffii for industrial production.

|
Scooped by
?
December 18, 6:37 PM
|
The detection of small molecules (e.g., pesticides, pollutants, antibiotics, drug residues) is critical for human health and environmental surveys as well as for many industrial applications. Fluorogenic RNA-based biosensors (FRBs) are aptamer-based tools specifically reporting on the presence of such targets by converting a specific binding event into fluorescence emission, thus making them of particular interest for on-site sensing applications. However, new sensor aptamers suited for FRB development are tedious to develop. In this study, we show that Capture-SELEX used in tandem with our microfluidic-assisted screening technology (μIVC) accelerates de novo FRB development by reprogramming the specificity of an existing sensor aptamer, followed by fluorescence-based functional selection. Furthermore, we show that these new sensor aptamers can be repurposed into functional biotechnological tools, such as gene-regulating aptazymes, thus demonstrating the versatility of our pipeline for the development of sensor aptamers adapted for various applications.
|
 Your new post is loading...
Your new post is loading...