 Your new post is loading...

|
Scooped by
?
Today, 6:27 PM
|
Overlapping genes allow multiple proteins to be encoded from a single DNA sequence, including convergent (antisense; tail-to-tail) orientations across three reading frames (phases 0, 1, and 2), with phase 1 most frequently observed in nature. Designing such overlaps is challenging due to codon degeneracy, phase-specific biases, and the need to preserve structural integrity for both proteins. Here, a purpose-built transformer encoder is introduced, trained on a balanced synthetic dataset of convergent overlaps spanning diverse prokaryotic genomes and GC contents. Controlled amino acid substitutions were incorporated during training to enhance model generalization, particularly for phase 1 overlaps. At inference, Monte Carlo dropout enabled uncertainty-aware sampling of synonymous codon solutions, which were iteratively refined using a windowed, multi-objective optimization framework. Candidate overlaps were scored using composite weighting across secondary structure preservation, substitution similarity, alignment identity, and ESM-2 contact map similarity, with SSIM applied as a rapid proxy for structural fidelity. This approach generated convergent overlaps across all phases, with phase 1 showing the highest success rates. Optimization trajectories revealed distinct dynamics, with secondary structure preservation steadily increasing despite its lower weight. External validation using SwissProt proteins stratified by AlphaFold2 pLDDT confidence supported generalization to proteins with differing rigidity, yielding high secondary structure preservation in silico. These results demonstrate that transformer models trained directly at the nucleotide level, when coupled with uncertainty-aware inference and lightweight structural proxies, can support the computational design of synthetic overlapping genes without requiring full structural prediction. This framework offers a scalable path for phase-specific, codon-aware overlap design under realistic constraints.

|
Scooped by
?
Today, 3:33 PM
|
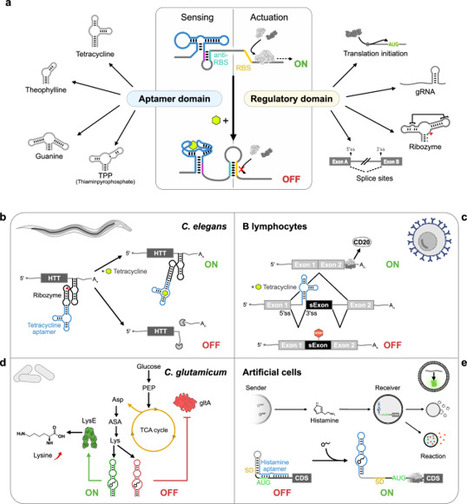
Synthetic riboswitches have undergone great development in the past decade, evolving into valuable regulatory tools. Operating entirely at the RNA level and independently of auxiliary proteins, they offer a promising alternative to protein-based systems such as TetON/OFF or CRISPR-Cas. As compact, modular RNA elements they unite sensing and regulatory functions within a single molecule, giving them the advantages of high modularity, portability and low metabolic burden. Here, we explore the unique features of synthetic riboswitches, highlight key applications, assess current bottlenecks and limitations and put them in context with emerging solutions, to emphasise the potential of synthetic riboswitches. Synthetic riboswitches are a burgeoning regulatory tool in the field of molecular biology. Here, the authors explore the unique features of synthetic riboswitches, highlight key applications, assess current bottlenecks and limitations and put them in context with emerging solutions.

|
Scooped by
?
Today, 3:22 PM
|
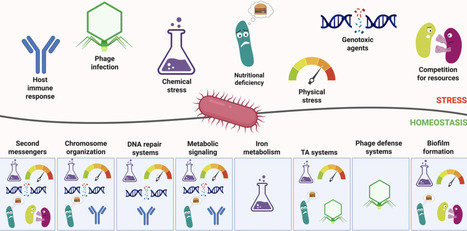
Bacteria have been generally greatly overlooked in the aspect of intra- and extra-cellular homeostasis, and yet, since they have evolved intricate processes and mechanisms allowing them not only to stay alive but also thrive in favorable and unfavorable environments alike, they should be considered as a close-to-ideal example of single-cell homeostasis. The bacterial responses aimed at maintaining homeostasis, while adjusting and reacting smoothly and swiftly to any changes inside and outside the cell, involve complex transcriptional networks regulated by second messengers and DNA topology, but also influenced by the presence of prophages and toxin-antitoxin systems. Their adjustment to nutrient availability also involves homeostasis in energy-related processes, such as central carbon metabolism, and crucial ion acquisition, e.g., iron. The genome stability, which is indispensable to maintain a given organisms’ functions, is achieved by control of DNA replication and repair. Furthermore, bacteria can form multicellular structures (biofilms), where homeostasis is achieved at several different levels and provides bacteria with higher chances of survival and colonization of new niches and locations. Precise correlation between the above-mentioned cellular processes makes bacteria highly intriguing objects of studies. Homeostasis is the most important basis of their life-style flexibility, thus understanding of these processes is indispensable for both: the basic and applied sciences. For example, understanding how chromosomal architecture and DNA topology coordinate global gene expression is essential for optimizing strain engineering and synthetic biology applications. Moreover, bacterial homeostasis regulatory processes can be employed as targets for antibacterial agents and prospective therapies.

|
Scooped by
?
Today, 2:16 PM
|
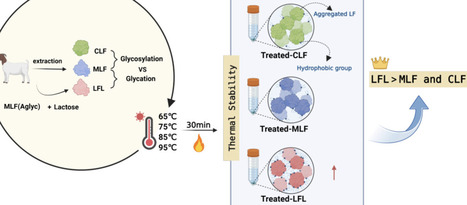
Lactoferrin (LF) is a heat-sensitive, iron-binding globular glycoprotein. Glycosylation and glycation can significantly alter its three-dimensional structure, spatial conformation, and functional properties, thereby affecting its thermal stability. This study compared the thermal stability of glycosylated LF from caprine colostrum (CLF) and mature milk (MLF) with that of glycated LF from lactosylated LF (LFL). During the heating process, both CLF and MLF exhibited heat-induced aggregation in SDS-PAGE, the holo-peak of CLF was higher than that of MLF after heat treatment, and the thermal transition temperature of CLF (95 °C) was higher than that of MLF (85 °C), suggesting glycosylation plays a role in the heat stability of LF. Compared to MLF, LFL exhibited less thermal aggregation and greater retention of secondary structure. In addition, the LFL showed more rod-shaped proteins in SEM, indicating that the LFL had improved thermal stability. This study reveals the potential effects of glycosylation and glycation in enhancing the thermal stability of LF.

|
Scooped by
?
Today, 1:49 PM
|
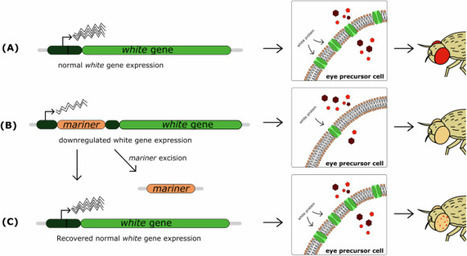
mariner is a transposable element of the Tc1/mariner superfamily that is widely distributed in various species. It was discovered in Drosophila mauritiana owing to a white-peach eye color mutation, and since then it has been used as a research tool in many systems and species. mariner element mobilization consists of cut-and-paste transposon excision and insertion. Here, apart from giving a historical overview of the discovery, distribution, and classification of mariner elements, we address the factors responsible for their particularly high somatic mobilization activity, with a focus on stress responses. We also address the usage of mariner transposases as research tools and how somatic mobilization can currently be detected.

|
Scooped by
?
Today, 1:35 PM
|
Ribonuclease P (RNase P), a ribozyme conserved across all domains of life, is involved in the tRNA 5′ maturation. RNase P recognizes precursor tRNA based on its structure, not the tRNA sequence. This feature had been exploited to engineer RNase P to selectively target and cleave any RNA as a gene inactivation strategy. Of these, a strategy called M1GS involves tethering an Escherichia coli M1 RNA to a short stretch of guide sequence (GS), which is complementary to the RNA targeted for cleavage. Despite its simplicity and versatility, M1GS tool appears to be underutilized compared to other gene inactivation strategies. Perhaps one of the reasons is that employment of the M1GS strategy requires prior knowledge about the requirements of the M1GS target sites. To facilitate its broader use, we have developed a Python script-based user-friendly bioinformatic tool built based on the requirements of M1GS to predict its target sites for any given RNA (using either DNA or RNA sequence as an input). In this study, we first demonstrate the utility of the bioinformatic tool in predicting M1GS target sites for human 28S rRNA and then we show that the customized M1GS-mediated downregulation of 28S rRNA in a human cancer cell line. We further validate the bioinformatic tool by predicting M1GS target sites for two previously targeted RNAs. Lastly, we discuss the utility of M1GS ribozyme-mediated rRNA downregulation as a potential anticancer modality in cancers where rRNAs are upregulated.

|
Scooped by
?
Today, 11:00 AM
|
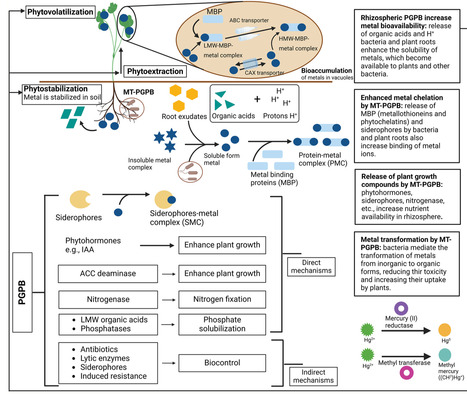
Heavy metal (HM) contamination in agricultural soils threatens food security, soil health, and human well-being. While phytoremediation offers a sustainable alternative to conventional remediation methods, its efficiency remains limited. Recent advances in artificial intelligence (AI), machine learning (ML), and multiomics technologies (genomics, proteomics, metabolomics) provide transformative opportunities to overcome these limitations. This review highlights the integration of AI-driven models with multiomics data to optimize phytoremediation strategies. AI enables the prediction of plant–microbe interactions, selection of plant growth-promoting bacteria (PGPB), and modeling of metal transporter dynamics, thereby enhancing crop tolerance and metal accumulation. By mining large-scale omics datasets, AI can also identify critical pathways for detoxification and guide precision engineering of plants and microbes. The convergence of AI, ML, and multi-omics technologies represents a transformative approach to solving the challenge of heavy metal pollution in soils. This integrated framework not only accelerates the development of metal-resistant crops but also paves the way for a new era of precision remediation, where tailored, data-driven solutions could revolutionize soil decontamination and lead to more sustainable and resilient agricultural practices.

|
Scooped by
?
Today, 10:42 AM
|
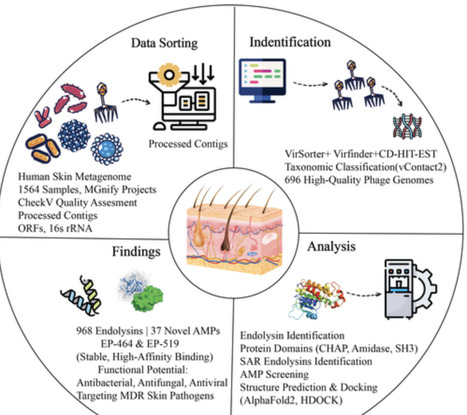
The global rise of antibiotic-resistant pathogens has intensified the search for alternative therapeutics. Bacteriophage-derived endolysins are emerging as promising candidates. They exhibit strong potential due to their target specificity, rapid bactericidal action, and low tendency to induce bacterial resistance. This study presents a comprehensive metagenomic analysis of the human skin phageome using 1564 samples from 10 metagenomic projects. Our analysis led to the classification of 696 phage genomes into clusters and singletons. These genomes displayed considerable variation in size, GC content (average 56%), and coding efficiency (72%). A total of 968 endolysins were identified, including 75 SAR variants, with diverse domain architectures such as CHAP, Amidase, and SH3, suggesting host-specific adaptations. Notably, we identified 37 previously unreported endolysin-derived antimicrobial peptides (AMPs), several of which exhibited nontoxic, antifungal, and antiviral properties. Molecular dynamics and docking studies revealed strong binding affinity and stability of peptides EP-464 and EP-519 to key virulence factors, including Staphylococcus epidermidis autolysin (PDB: 4EPC), beta-lactamase VIM-2 (PDB: 5O7N), and AHL synthase LasI (PDB: 1RO5). These interactions suggest potential for disrupting bacterial virulence, resistance mechanisms, and quorum sensing. This study provides the first large-scale functional characterization of the human skin phageome focused on therapeutic endolysins and their novel AMP derivatives, offering promising candidates for the development of next-generation antimicrobial agents. However, further experimental validation is essential to assess their clinical efficacy in treating skin-related infections.

|
Scooped by
?
Today, 10:33 AM
|
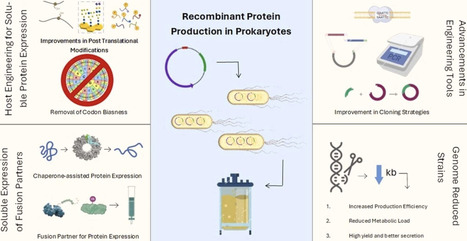
Recombinant protein production in prokaryotic systems remains a major topic in biotechnology because of their rapid growth, cost-effectiveness, and ease of genetic manipulation. However, the production of functionally active proteins still faces significant challenges due to folding failures, insolubility, and the lack of the capability of most prokaryotes for complex post-translational processing. This review dwells into both traditional and emerging strategies for optimizing recombinant protein expression in various prokaryotic systems. It also highlights recent advances in genetic engineering and synthetic biology for expanding the toolkit available for protein production, which include refined expression vectors, engineered hosts with improved folding capabilities, and high-throughput screening platforms. Additionally, it provides a thorough discussion of how to optimize heterologous expression using fusion tag approaches, codon bias elimination, promoter and ribosome binding site (RBS) engineering, and chaperone-assisted folding. This review explores diverse prokaryotic expression systems that offer unique advantages for heterologous expression that extend far beyond the limitations of traditional hosts. Additionally, this review also emphasizes the need for the selection of the right expression system and optimizing conditions to fulfill the increasing demands for recombinant protein production in various fields.

|
Scooped by
?
Today, 10:04 AM
|
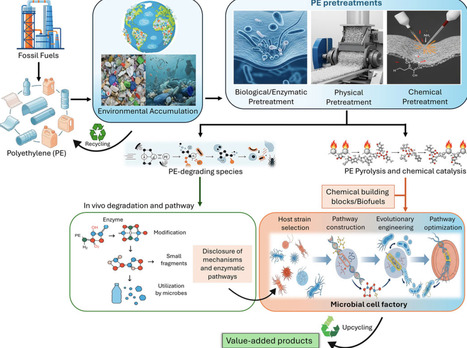
Polyethylene (PE) is one of the widely utilized plastics globally, valued for its durability but unsustainable due to its resistance to biodegradation in a natural environment, leading to severe environmental accumulation. Recent studies have identified microorganisms, insects, and potential PE-degrading enzymes (PEases) capable of breaking down PE, suggesting a possible route for biorecycling. However, research in this area remains in its early stages, with limited understanding of the enzymatic mechanisms involved and the degradation products formed. A major barrier lies in the chemically inert nature of PE’s carbon–carbon and carbon–hydrogen bonds, which makes enzymatic degradation particularly challenging and unlikely to occur through a single enzyme. Overcoming these limitations requires the discovery and engineering of complex enzymatic pathways, supported by emerging tools such as omics technologies, structure-guided design, and computer-aided enzyme engineering. In parallel, the biotechnological upcycling of PE waste into value-added chemicals, by first breaking down PE into smaller products and then using them as microbial feedstocks, holds significant potential but is currently underexplored. To date, polyhydroxyalkanoate (PHA) remains the most studied PE waste upcycled biopolymer product, with only a few other studies showing production of diacids, protein, wax esters, and lipids. This highlights the need for expanded research into microbial metabolism and metabolic engineering to enable more diverse and efficient PE waste bioconversion routes. This review summarizes the current state as an integrated effort for biorecycling of PE, including PE pretreatment technologies, enzymatic PE degradation, microbial PE degradation, and PE upcycling into value-added chemicals via metabolic engineering. This review also highlights key scientific challenges and outlines future directions for PE degradation and transforming PE waste into valuable and sustainable products.

|
Scooped by
?
November 9, 11:52 PM
|
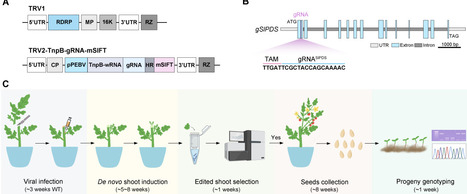
Genome editing has emerged as a powerful tool for crop breeding. However, most commonly used approaches rely on tissue culture and the use of transgenic materials, which are time-consuming and labor-intensive, and often exhibit strong genotype dependency. Here we utilized tobacco rattle virus (TRV)-mediated ISYmu1 genome editing, coupled with in planta shoot regeneration, to achieve somatic and heritable genome editing in different tomato cultivars. By targeting the SlPDS gene, we successfully generated virus- and transgene-free homozygous mutant progeny in one generation. We further verified the method by generating somatic edits in SlDA1, a gene with potential agronomic relevance. Given the wide host range of TRV and its compatibility with in planta shoot regeneration, our system could be broadly applicable for rapid, non-transgenic and heritable genome editing, thereby advancing functional genomics and crop improvement.

|
Scooped by
?
November 9, 11:22 PM
|
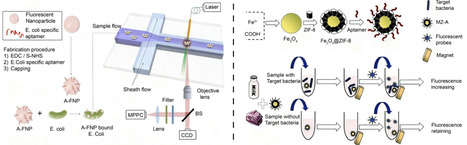
Foodborne pathogen contamination is a major global public health issue, highlighting the need for rapid, sensitive, and portable detection technologies. Conventional methods such as microbial culture, polymerase chain reaction, and enzyme-linked immunosorbent assay are limited by complexity, low sensitivity, and high cost, restricting on-site use. Functional nucleic acids (FNAs), including aptamers and DNAzymes, have emerged as versatile molecular recognition elements for next-generation biosensors. Their programmability, stability, and specificity enable reliable real-time food monitoring. Moreover, nucleic acid-based signal amplification methods greatly enhance detection sensitivity. This review summarizes recent advances in FNA-based biosensors for foodborne pathogen detection, covering screening strategies like systematic evolution of ligands by exponential enrichment, sensing mechanisms with various readouts (fluorescence, colorimetry, electrochemistry), and innovative amplification strategies for improving practical application in food safety monitoring. Representative biosensor designs, mechanisms, and performances are highlighted. Finally, existing challenges and future perspectives for enhancing detection accuracy are discussed. FNA-driven biosensors hold great promise for preventing foodborne diseases and advancing portable, high-sensitivity detection platforms.

|
Scooped by
?
November 9, 11:13 PM
|
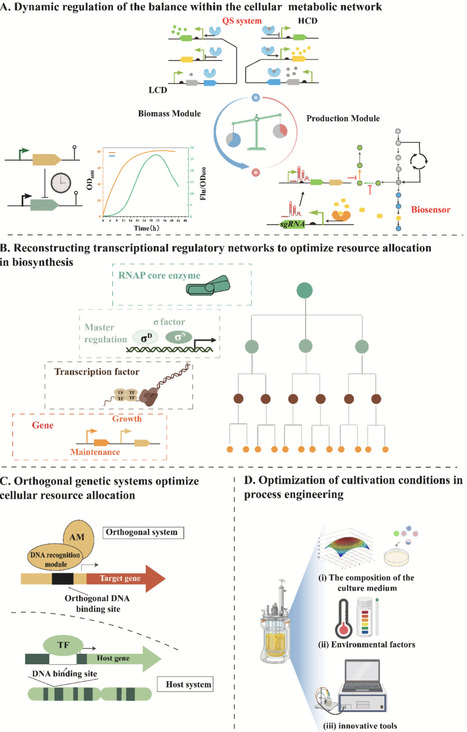
Cellular activity serves as a critical determinant of growth, metabolism, and production efficiency in microbial cell factories. However, industrial-scale microbial fermentation is often limited by reduced cellular activity due to metabolite toxicity, metabolic burden, and environmental stress, which constrain productivity and compromise target compound synthesis. This review systematically explores contemporary strategies to enhance cellular activity, focusing on three aspects: (i) alleviating toxicity from substrates, intermediates, and products; (ii) optimizing cellular resource allocation and cofactor synthesis to reduce metabolic burden; and (iii) developing adaptive evolution techniques, screening tolerance targets, engineering cell membranes, and employing exogenous protective agents to bolster cell resistance to environmental stress. Recent advances addressing these challenges are summarized, aiming to enhance the production capacity and stability of microbial cell factories. Finally, we discuss persistent challenges and future research priorities in improving cellular activity.
|

|
Scooped by
?
Today, 3:40 PM
|
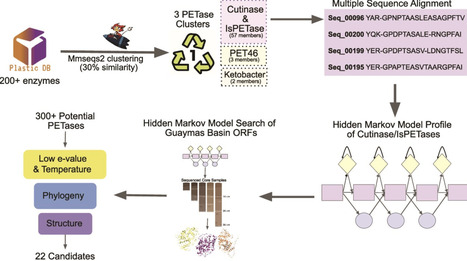
Polyethylene terephthalate (PET)-hydrolyzing enzymes (PETases) are a recently discovered enzyme class capable of plastic degradation. PETases are commonly identified in bacteria; however, pipelines for discovery are often biased to recover highly similar enzymes. Here, we searched metagenomic data from hydrothermally impacted deep sea sediments in the Guaymas Basin (Gulf of California) for PETases. A broad diversity of potential proteins were identified and 22 were selected based on their potential thermal stability and phylogenetic novelty. Heterologous expression and functional analysis of these candidate PETases revealed three candidates capable of depolymerizing PET or its byproducts. One is a PETase from a Bathyarchaeia archaeon (dubbed GuaPA, for Guaymas PETase Archaeal) and two bishydroxyethylene terephthalate-hydrolyzing enzymes (BHETases) from uncultured bacteria, Poribacteria, and Thermotogota. GuaPA is the first archaeal PETase discovered that is able to depolymerize PET films and originates from a specific enzyme class which has endowed it with predicted novel structural features. Within 48 h, GuaPA released ~3–5 mM of terephthalic acid and mono-(2-hydroxyethyl) terephthalate from low crystallinity PET. PET co-hydrolysis containing GuaPA and one of the newly discovered BHETases further improves the hydrolysis of untreated PET film by 68%. Genomic analysis of the PETase- and BHETase-encoding microorganisms reveals that they likely metabolize the products of enzymatic PET depolymerization, suggesting an ecological role in utilizing anthropogenic carbon sources. Our analysis reveals a previously uncharacterized ability of these uncultured microorganisms to catabolize PET, suggesting that the deep ocean is a potential reservoir of biocatalysts for the depolymerization of plastic waste.

|
Scooped by
?
Today, 3:28 PM
|
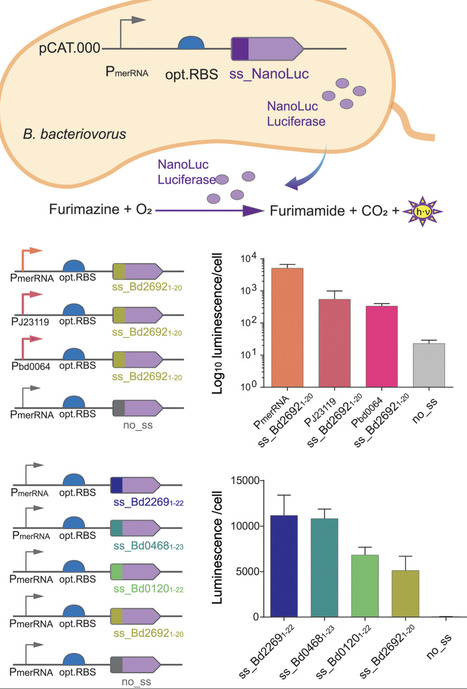
The predatory bacterium Bdellovibrio bacteriovorus kills and consumes other bacteria, thrives in diverse environments and holds great potential to address major challenges in medicine, agriculture, and biotechnology. As a bacterial predator it represents an alternative to traditional antimicrobial strategies to combat multidrug-resistant bacterial pathogens and prevent food waste, while the multitude of predatory enzymes it produces have potential for biotechnological applications. However, while a limited set of genetic tools exist, the lack of secretion assays and fine-tuning of secretion constrain both fundamental studies and bioengineering of B. bacteriovorus. Here, we present a molecular toolbox for B. bacteriovorus by systematically tuning gene expression and secretion of a reporter protein. Building on functional native and synthetic promoters from the Anderson library with varying expression levels of fluorescent reporter protein mScarletI3, we evaluated different ribosomal binding sites (RBS) to fine-tune gene expression. To examine secretion, we established a novel protocol to quantify extracellular release of a Nanoluc luciferase reporter protein in B. bacteriovorus using different native Sec-dependent signal sequences. We anticipate that the newly developed genetic toolkit and techniques will advance research on this fundamental predator-prey system, laying the foundation for its broader application and future bioengineering efforts. This work will pave the way for tailored applications of B. bacteriovorus in microbial ecology, agriculture, biotechnology, and medicine.

|
Scooped by
?
Today, 3:17 PM
|
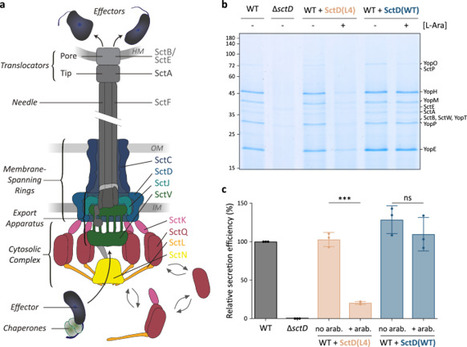
Major bacterial pathogens manipulate eukaryotic target cells by injecting effector proteins through type III secretion systems (T3SS). Recent in situ observations revealed that these large molecular machines, often referred to as injectisomes, are remarkably dynamic and adaptive entities, with the cytosolic T3SS components forming a mobile network that recruits effectors to the export machinery. In contrast to these soluble components, the transmembrane rings anchoring the injectisome are stably associated – with one exception. Using functional assays, live cell microscopy, and photobleaching experiments, we found that SctD, which constitutes the inner membrane ring of the T3SS, exchanges subunits in secreting injectisomes in Yersinia enterocolitica. To elucidate the biological significance of this unexpected dynamic behavior of a key structural component, we investigated its role in the assembly and function of the T3SS. Using engineered SctD variants whose exchange rate can be modulated, we found that exchange supports the integration of export apparatus components into assembled membrane rings and efficient secretion of effectors. Our findings uncover a new aspect of the molecular function and regulation of the T3SS, which may apply to other secretion systems and molecular machines. Bacteria use the type III secretion system (T3SS) to inject proteins into target cells. Here, Brianceau et al. show that a core structural component, SctD, exchanges between a T3SS-bound and a freely diffusing state in Yersinia bacteria, and that this exchange is required for assembly and function of the T3SS.

|
Scooped by
?
Today, 2:05 PM
|
Pan-genome analysis is a crucial method for studying genomic dynamics. By creating pan-genome maps for prokaryotic organisms, we can gain valuable insights into their genetic diversity and ecological adaptability. However, current analytical methods often struggle to balance accuracy and computational efficiency, and they tend to provide primarily qualitative results. This study introduces PGAP2, an integrated software package that simplifies various processes, including data quality control, pan-genome analysis, and result visualization. PGAP2 facilitates the rapid and accurate identification of orthologous and paralogous genes by employing fine-grained feature analysis within constrained regions. Our systematic evaluation with simulated and gold-standard datasets demonstrates that PGAP2 is more precise, robust, and scalable than state-of-the-art tools for large-scale pan-genome data. Furthermore, PGAP2 introduces four quantitative parameters derived from the distances between or within clusters, enabling detailed characterization of homology clusters. Finally, we validate our quantitative findings by applying PGAP2 to construct a pan-genomic profile of 2794 zoonotic Streptococcus suis strains. This analysis offers new insights into the genetic diversity of S. suis, thereby enhancing our understanding of its genomic structure. PGAP2 is freely available at https://github.com/bucongfan/PGAP2 . Prokaryotic pan-genome analysis is crucial for understanding microbial diversity, however current analytical methods often struggle to balance accuracy and computational efficiency. Here the authors present a more precise, robust and scalable toolkit for large-scale pan genome analysis.

|
Scooped by
?
Today, 1:40 PM
|
Antibiotic susceptibility tests (ASTs) often fail to predict treatment outcomes because they do not account for biofilm-specific tolerance mechanisms. In the present study, we explored alternative approaches to predict tobramycin susceptibility of Pseudomonas aeruginosa biofilms that were experimentally evolved in physiologically relevant conditions. To this end, we used four analytical methods – whole-genome sequencing (WGS), matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), isothermal microcalorimetry (IMC) and multi-excitation Raman spectroscopy (MX-Raman). Machine learning models were trained on data outputs from these methods to predict tobramycin susceptibility of our evolved strains and validated with a collection of clinical isolates. For minimal inhibitory concentration (MIC) predictions of the evolved strains, the highest accuracy was achieved with MALDI-TOF MS (97.83%), while for biofilm prevention concentration (BPC) predictions, Raman spectroscopy performed best with an accuracy of 80.43%. Overall, all analytical methods demonstrated comparable predictive performance, showing their potential for improving biofilm AST.

|
Scooped by
?
Today, 12:48 PM
|
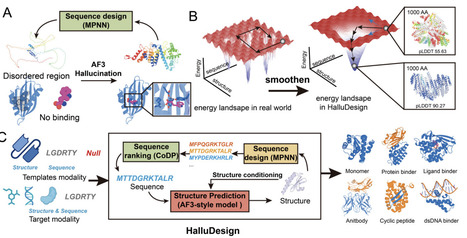
Deep learning has revolutionized biomolecular modeling, enabling the prediction of diverse structures with atomic accuracy. However, leveraging the atomic-level precision of the structure prediction model for de novo design remains challenging. Here, we present HalluDesign, a general all-atom framework for protein optimization and de novo design, which iteratively update protein structure and sequence. HalluDesign harnesses the inherent hallucination capabilities of AlphaFold3-style structure prediction models and enables fine-tune free, forward-pass only sequence-structure co-optimization. Structure conditioning at different noise level in the structure prediction stage allows precise control over the sampling space, facilitating tasks from local and global protein optimization to de novo design. We demonstrate the versatility of this approach by optimizing suboptimal structures, rescuing previously unsuccessful designs, designing new biomolecular interactions and generating new protein structures from scratch. Experimental characterization of binder design spanning small molecule, metal ion, protein, and antibody design of phosphorylation-specific peptide revealed high design success rates and excellent structural accuracy. Together, our comprehensive computational and experimental results highlight the broad utility of this framework. We anticipate that HalluDesign will further unlock the modeling and design potential of AlphaFold3-like models, enabling the systematic creation of complex proteins for a wide range of biotechnological applications.

|
Scooped by
?
Today, 10:52 AM
|
Nature has spent billions of years evolving the most efficient and effective solutions to complex problems, from navigation and energy harvesting to visual processing and biodegradation. Bioinspired engineering draws on these strategies to design adaptive, efficient and sustainable technologies, particularly in fields such as robotics, materials science and medical device engineering. robots typically have difficulty transversing uneven or unstable terrain, and even the most efficient bipedal robots fall behind humans in terms of energy usage. In a Review in this issue, Barbara Mazzolai and team analyse the cost of transport for different animals, plants and robotic systems to identify energy-efficient movement strategies that can inform robot design. In a Comment in this issue, Weimin Zhang and colleagues outline the design of robots that mimic these behaviours by integrating multiple sensors and using neuromorphic chips or algorithms trained on animal patterns to process sensory information for efficient navigation.

|
Scooped by
?
Today, 10:38 AM
|
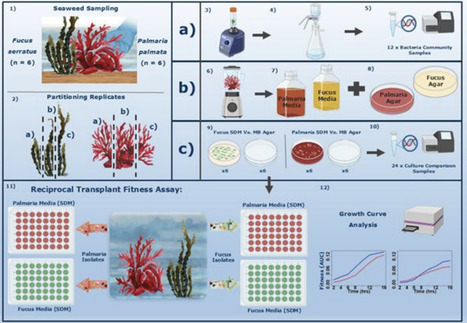
Seaweed microbiomes are diverse and frequently species-specific. By actively attracting and repelling settling bacteria through exuded metabolites, seaweeds are thought to exert a strong selective pressure on their microbiomes. However, to what degree seaweed-associated bacteria are adapted to their host has received little attention. Here, we retrieve cultivable seaweed bacterial communities from Palmaria palmata (Dulse) and Fucus serratus (Serrated Wrack) and use reciprocal transplant experiments to test whether bacterial isolates have the greatest fitness on their host seaweed species. We used agar derived from host seaweed extracts for bacterial isolation, which was found to be superior to a generic marine agar formulation based on both 16S rRNA gene amplicon alpha- and beta-diversity comparisons to uncultured samples. We then demonstrate that bacterial isolates from both seaweed species exhibit higher fitness in media derived from their native host compared to a non-native host. Although epibacterial fitness varied between hosts, bacterial isolates on average outperformed non-native counterparts in their native environment. By integrating amplicon sequencing with laboratory experiments, we demonstrate that bacteria are locally adapted to their seaweed host species. These findings contribute to the growing body of research exploring the evolutionary and ecological drivers that shape bacterial communities, with implications for ecosystem management, disease control and microbial biotechnology.

|
Scooped by
?
Today, 10:24 AM
|
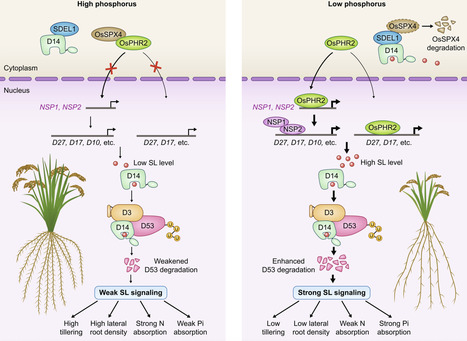
Strigolactones (SLs) are intriguing phytohormones that play essential roles in branch or tiller development and adaptation to nutrient availability. Tillering control is a means of improving the grain yield of cereal crops, particularly under nutrient-limited conditions. Recent research has provided new insights into the activation, termination and regulatory mechanisms of SL perception, as well as exciting insights into the low nitrogen (LN)-triggered phosphorylation of SL receptors, which is crucial for the tillering response to fluctuations in nitrogen availability. Low phosphorus (LP) induces accumulation of SLs, which inhibit tillering and facilitate the balance of nitrogen and phosphorus through the SL signaling pathway. Current understanding of SL-mediated nutrient responses offers promising avenues for molecular breeding strategies aimed at improving crop yield and resource use efficiency.

|
Scooped by
?
November 9, 11:54 PM
|
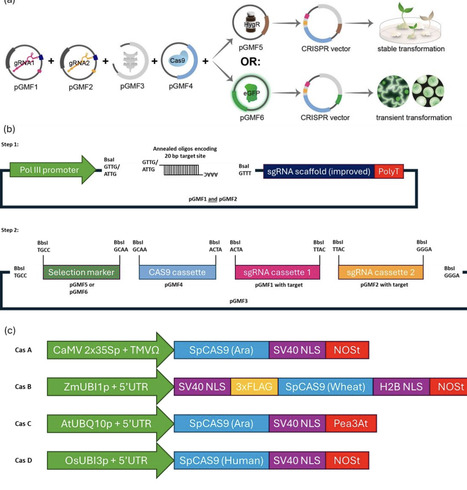
Gene editing, especially the CRISPR/Cas9 system, has revolutionized trait development in crops. However, large parts of the world are missing out on applying CRISPR in planta. There is an obvious lack of gene editing applications in locally relevant crops in the Global South which tend to be neglected by mainstream agricultural research and development. Access barriers to these new breeding technologies need to be removed to allow the potential impact of these technologies on food security to happen. Here, we present the ENABLE® Gene Editing in planta toolkit, a minimal molecular toolbox allowing users to create a CRISPR knockout vector for transient or stable plant transformation in two simple cloning steps. We validate the toolkit in rice (Oryza sativa) protoplasts and in Arabidopsis thaliana plants. The ENABLE® kit is designed to be utilized specifically by users in the Global South who are new to CRISPR technology by providing a simple workflow, extensive accompanying protocols as well as options for low cost methods for cloning verification and gene editing verification in planta. We hope that our toolkit helps bridge the gap between the recent biotechnological advancements in plant breeding that high income countries can access and the lack of those technologies in low and middle income countries.

|
Scooped by
?
November 9, 11:33 PM
|
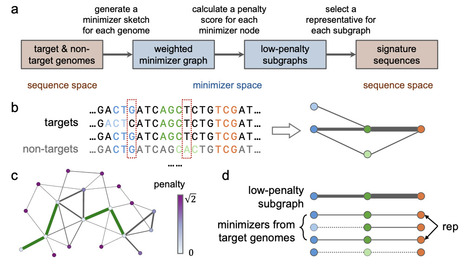
PCR enables rapid, cost-effective diagnostics but requires prior identification of genomic regions that allow sensitive and specific identification of target microbial groups, herein referred to as microbial signature sequences. We introduce Seqwin, an open-source framework designed to automate microbial genome signature discovery. Tens of thousands of microbial genomes are now available, limiting the application of existing manual and automated approaches for identifying signatures. Modern approaches that are capable of leveraging all available microbial genomes will ensure sensitive and accurate DNA signatures identification and enable robust pathogen detection for clinical, environmental, and public health applications. Seqwin builds weighted pan-genome minimizer graphs and uses a traversal algorithm to identify signature sequences that occur frequently in target genomes but remain rare in non-targets. Unlike earlier tools that depend on strict presence or absence of sequences, Seqwin accommodates natural sequence variation and scales to very large genome collections. When applied to genomes from C. difficile, M. tuberculosis, and S. enterica, Seqwin recovered more high-quality signatures than alternative methods with lower computational burden. Seqwin analysis of nearly 15,000 S. enterica genomes yielded over 200 candidate signatures in less than 10 minutes. Seqwin provides an open-source solution for the long-standing need for scalable microbial signature discovery and diagnostic assay design. https://github.com/treangenlab/Seqwin

|
Scooped by
?
November 9, 11:19 PM
|
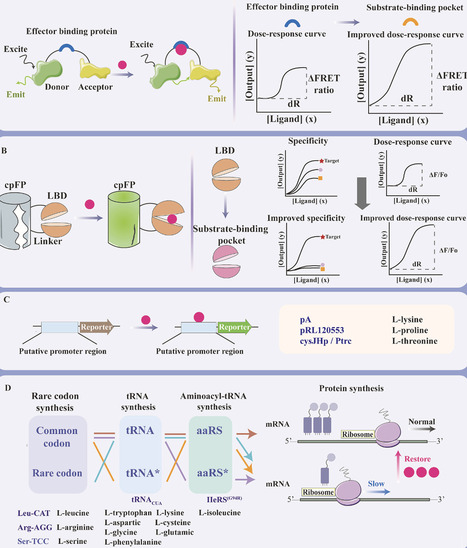
Amino acids and their derivatives play pivotal roles across diverse fields including biotechnology, pharmaceuticals, agriculture, and industrial manufacturing. The development of high-throughput screening methods for strains producing amino acids and their derivatives is crucial for both mining key enzymes and screening overproducers. This review systematically evaluates six classes of direct biosensors employed in the metabolic engineering of amino acid- or derivative-producing strains. These include biosensors based on transcription factors, riboswitches, Förster resonance energy transfer FRET, circularly permuted fluorescent proteins, compound-inducible putative promoter regions, and protein translation elements. Their operational principles and recent advances in rational design, performance optimization, and practical implementation are critically analyzed. In addition, a systematic analysis of four categories of indirect biosensing strategies for the screening or regulation of amino acid- or derivative-producing strains is provided. These strategies target universal metabolic precursors, pathway-specific precursors, enzymatically transformed downstream metabolites, or competitive intermediates in branched pathways. Then, the design strategies, performance optimization methods, and practical implementation challenges of the existing biosensors are compared, which are accompanied by the discussion of the key parameters that are optimal for the biosensors applied in metabolic engineering. This work will facilitate the development of biosensors for metabolites that currently lack biosensing systems, and promote the innovation of the existing biosensors. These developments are expected to support efficient and sustainable production of amino acid-related compounds and other high-value metabolites.
|
 Your new post is loading...
Your new post is loading...














Following this, molecular docking of the seven most stable AMPs with the selected protein target was performed using the HDOCK (http://hdock.phys.hust.edu.cn/) server. The target proteins included the Staphylococcus epidermidis virulence factor autolysin (PDB: 4EPC) (Zoll et al. 2012), Beta-lactamase VIM-2 (PDB: 5O7N), and the AHL Synthase LasI (PDB: 1RO5)