 Your new post is loading...

|
Scooped by
?
Today, 1:08 AM
|
Lignin, a natural and renewable aromatic polymer, serves as a plentiful source of aromatic building blocks for biomanufacturing. However, despite its potential, the bioconversion efficiency of lignin remains limited due to its inherently complex structure and the constraints of traditional metabolic engineering approaches. Synthetic biology-guided metabolic regulation offers a solution by coordinating intracellular resource allocation in ligninolytic strains, endowing their adaptation for industrial applications and promoting the sustainability of lignin-based bioeconomy. This review provides a comprehensive overview of multiscale metabolic regulation, including critical enzymes involved in biotransformation, metabolic pathway networks, genome-phenotype and learning prediction. It highlights the significant roles and potential benefits of emerging cutting-edge technologies in advancing lignin valorization. Overall, synthetic biology-guided metabolic regulation has demonstrated its power in balancing the metabolic fluxes of ligninolytic strains, ensuring the continued vitality in future development.

|
Scooped by
?
Today, 12:56 AM
|
Methane emissions from ruminants represent a major contributor to global greenhouse gases (GHGs), posing challenges for sustainable agriculture and climate change mitigation. Recent advances in CRISPR-Cas-based gene editing offer transformative approaches to address this issue by targeting both forage crops and rumen methanogens. Enhancing lipid content and secondary metabolites in forage crops can suppress methanogenesis and improve feed efficiency, while precise genome editing in methanogenic Archaea can disrupt pathways critical to methane production. However, challenges remain regarding delivery methods, gene targeting specificity, ecological impacts, and regulatory acceptance. In this review, we explore current progress, identify key knowledge gaps, and highlight the need for interdisciplinary strategies that integrate biotechnology, synthetic biology, and regulatory frameworks to develop effective and scalable methane mitigation solutions.

|
Scooped by
?
Today, 12:40 AM
|
Lignocellulosic wine grape waste (WGW) is a cheap medium for Escherichia coli growth and H2 production. The current study investigated the effect of initial redox potential (oxidation–reduction potential (ORP)) on the growth, H2 production, ORP kinetics, and current generation of the E. coli BW25113 parental and a mutant strain optimized for hydrogen evolution when fermenting WGW (40 g L−1) hydrolysate. Bacteria were cultivated anaerobically on pre-treated WGW hydrolysate with dilutions ranging from undiluted to fourfold dilution, at pH 7.5. Notably, a twofold diluted medium, with pH adjustment using K2HPO4, exhibited reduced acidification, prolonged H2 production, and enhanced biomass formation (OD600, 1.5). The addition of the redox reagent DL-dithiothreitol (DTT) was found to positively influence the H2 production of both the E. coli BW25113 parental and mutant strains. H2 production started after 24 h of growth, reaching a maximum yield of 5.10 ± 0.02 mmol/L in the wild type and 5.3 ± 0.02 mmol/L in the septuple mutant strain, persisting until the end of the stationary growth phase. The introduction of 3 mM DTT induced H2 production from the early-exponential phase, indicating that reducing conditions enhanced H2 production. Furthermore, we assessed the efficacy of using intact E. coli cells (1.5 mg cell dry weight) as anode catalyst in a bio-electrochemical fuel-cell system. Whole cells of the septuple mutant grown under reduced ORP conditions yielded the highest electrical potential, reaching up to 0.7 V. The results highlight the potential of modifying medium buffering capacity and ORP as a tool to improve biomass yield and H2 production during growth on WGW for biotechnological biocatalyst applications.

|
Scooped by
?
Today, 12:30 AM
|
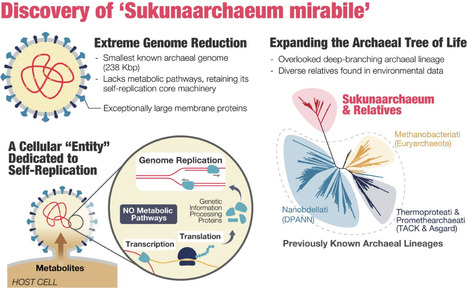
Defining the minimal genetic requirements for cellular life remains a fundamental question in biology. Genomic exploration continually reveals novel microbial lineages, often exhibiting extreme genome reduction, particularly within symbiotic relationships. Here, we report the discovery of Candidatus Sukunaarchaeum mirabile, a novel archaeon with an unprecedentedly small genome of only 238 kbp —less than half the size of the smallest previously known archaeal genome— from a dinoflagellate-associated microbial community. Phylogenetic analyses place Sukunaarchaeum as a deeply branching lineage within the tree of Archaea, representing a novel major branch distinct from established phyla. Environmental sequence data indicate that sequences closely related to Sukunaarchaeum form a diverse and previously overlooked clade in microbial surveys. Its genome is profoundly stripped-down, lacking virtually all recognizable metabolic pathways, and primarily encoding the machinery for its replicative core: DNA replication, transcription, and translation. This suggests an unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses. The discovery of Sukunaarchaeum pushes the conventional boundaries of cellular life and highlights the vast unexplored biological novelty within microbial interactions, suggesting that further exploration of symbiotic systems may reveal even more extraordinary life forms, reshaping our understanding of cellular evolution.

|
Scooped by
?
June 14, 11:55 PM
|
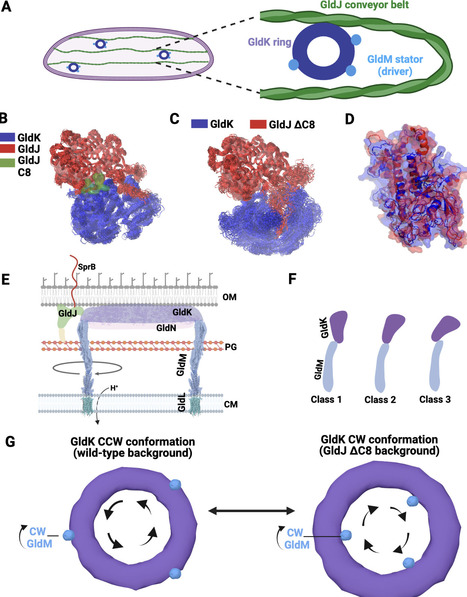
Many bacteria utilize the type 9 secretion system (T9SS) for gliding motility, surface colonization, and pathogenesis. This dual-function motor supports both gliding motility and protein secretion, where rotation of the T9SS plays a central role. Fueled by the energy of the stored proton motive force and transmitted through the torque of membrane-anchored stator units, the rotary T9SS propels an adhesin-coated conveyor belt along the bacterial outer membrane like a molecular snowmobile, thereby enabling gliding motion. However, the mechanisms controlling the rotational direction and gliding motility of T9SS remain elusive. Shedding light on this mechanism, we find that in the gliding bacterium Flavobacterium johnsoniae, deletion of the C-terminus of the conveyor belt-associated protein GldJ controls and, in fact, reverses the rotational direction of T9SS from counterclockwise (CCW) to clockwise (CW). This suggests that the interface between the conveyor belt-associated protein GldJ and the T9SS ring protein GldK plays an important role in controlling the directionality of T9SS, potentially by modulating its interaction with the stator complex GldLM, which drives motor rotation. Combined with MD simulation of the T9SS stator units GldLM, we suggest a “tri-component gearset” model where GldJ controls the rotational direction of its driver, the T9SS, thus providing adaptive sensory feedback to influence the motility of the gliding bacterium.

|
Scooped by
?
June 14, 11:38 PM
|
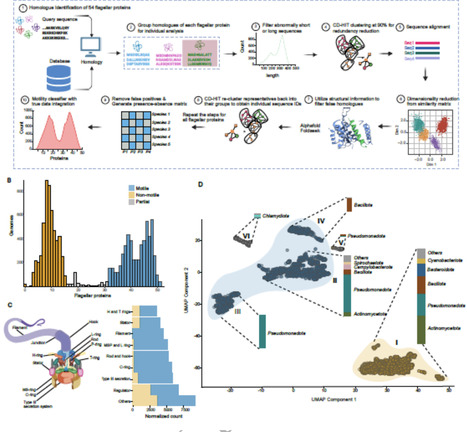
Bacterial swimming is mostly powered by the bacterial flagellar motor and the number of proteins involved in the flagellar motor can vary. Quantifying the proteins present in flagellar motors from a range of species delivers insight into how motility has changed throughout history and provides a platform for estimating from its genome whether a species is likely to be motile. We conducted sequence and structural homology searches for 54 flagellar pathway proteins across 11 365 bacterial genomes and developed a classifier with up to 95% accuracy that could predict whether a strain was motile or not. We then mapped the evolution of flagellar motility across the GTDB bacterial tree of life. We confirmed that the last common bacterial ancestor had flagellar motility and that the rate of loss of this motility was four-fold higher than the rate of gain. We showed that the presence of filament protein homologues was highly phylogenetically correlated with motility and that all species classified as motile contained at least one filament homologue. We calculated the rate of gain and loss for each flagellar protein and that the filament protein FliC was highly correlated with motility across the tree of life. We then measured the correlation of each flagellar motor protein with FliC and showed that the filament, rotor, and rod and hook proteins were all highly correlated with FliC, and thus with motility. We calculated the differential rates of gain and loss for each flagellar protein and quantified which genomes encoded for partial sets of flagellar proteins, indicating potential pathways by which motility could be lost. Overall, we show that filament, rod and hook and rotor proteins are conserved when flagellar motility is preserved and that the presence or absence of a FliC homologue is a good, simple predictor of whether or not a species has flagellar motility.

|
Scooped by
?
June 13, 4:54 PM
|
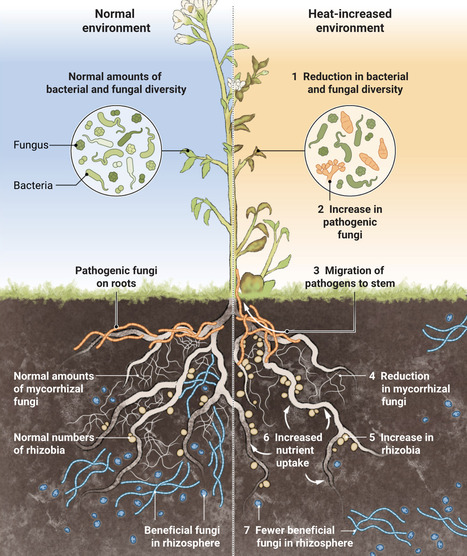
Rising temperatures change the structure and function of plant microbial communities.

|
Scooped by
?
June 13, 4:46 PM
|
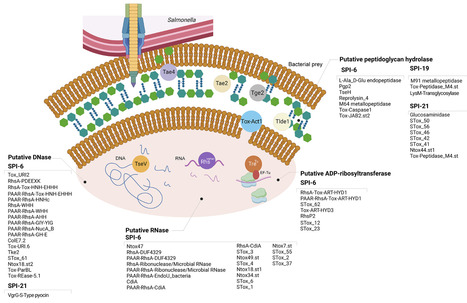
The Type VI Secretion System (T6SS) is a critical fitness and virulence factor of many Gram-negative bacteria. Five T6SS gene clusters have been described in Salmonella, each one encoded within different pathogenicity islands (i.e., SPI-6, SPI-19, SPI-20, SPI-21, and SPI-22). The events of gain and loss of these T6SS gene clusters have contributed to shape the genome evolution of different Salmonella serotypes. In addition, the differential distribution of T6SS and the ever-increasing repertoire of predicted effector proteins is likely to play an important role in the environmental fitness and pathogenic potential of different Salmonella serotypes. This review summarizes the current knowledge on the role played by T6SS in Salmonella biology, highlighting the major milestones in the field over the past two decades. We discuss the expanding repertoire of T6SS effector proteins identified to date and examine the current understanding of mechanisms controlling T6SS expression in Salmonella, focusing on host-derived cues and regulators involved. Finally, we provide a critical analysis of conflicting reports and suggest future directions for the research in the field. A better understanding of these processes could expand our knowledge of Salmonella biology, and the mechanisms behind how this versatile secretion system enables Salmonella to thrive in competitive microbial environments and contribute to host colonization.

|
Scooped by
?
June 13, 4:20 PM
|
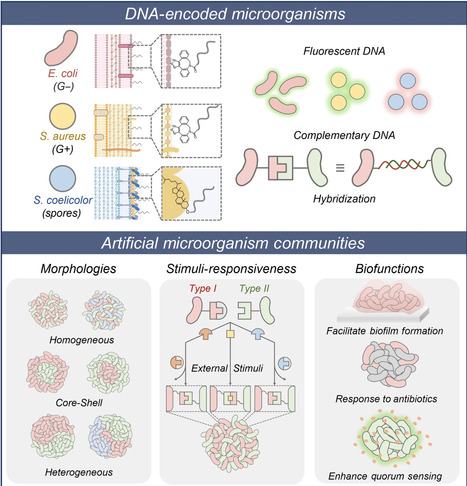
Programming microorganism adhesions to engineer multicellular microbial communities holds promise for synthetic biology and medicine. Current chemical and genetic engineering approaches often lack specificity or require engineered bacteria, making the design of responsive interactions challenging. Here, we demonstrate the use of functional DNA as programmable surface receptors to regulate the patterns and behaviors of microbial communities. Using metabolic labeling and hydrophobic insertion, we modified various microorganisms with DNA, including Gram-positive and Gram-negative bacteria, and dormant spores. By incorporating distinct sequences, we achieved precise spatial control of bi- and tricomponent microbial assemblies, forming diverse morphologies like core-shell and selective clusters. Stimuli-responsive clustering was successfully realized using aptamers, strand displacement, and reverse-Hoogsteen base pairing, with oligonucleotides or small molecules as exogenous cues. This work extends the use of functional DNA to control microbial interactions, enabling living communities with dynamic biofunctions, such as biofilm formation, antibiotic sensitivity, and quorum sensing, in response to biological triggers.

|
Scooped by
?
June 13, 3:25 PM
|
Root nodule symbiosis allows for plant acquisition of reactive nitrogen through fixation of atmospheric molecular dinitrogen by nitrogen-fixing bacteria. Nodulation is a complex trait, with diverse modes of bacterial infection and nodule morphologies across species, reflecting evolutionary adaptation. Understanding ancient forms of this trait may carry advantages for its current utilization, since basal states likely reflect the least complexity. In this review we focus on the evolution of nodule development, particularly on events that have led to increased complexity of this symbiosis in later adaptations. We hypothesize that the ancestral form of nodulation comprises of an evolutionary coupling of nutrient-dependent lateral root development with apoplastic intercellular bacterial growth, alongside the acquisition or evolution of an ancestral chitinaceous signaling molecule by the microbial symbiont. Uncovering the evolutionary adaptations underpinning the extant diversity of this trait allows for a better understanding of the simplest ancestral state.

|
Scooped by
?
June 13, 10:46 AM
|
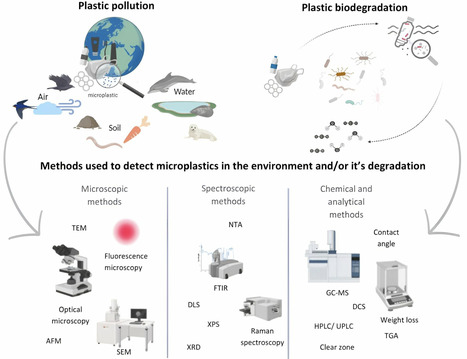
Environmental concerns about the widespread use of non-biodegradable plastic have generated interest in developing quick and effective methods to degrade synthetic polymers. With millions of tons of plastic waste generated annually, biodegradation by microorganisms presents a promising and eco-friendly solution. However, a bottleneck has arisen due to the lack of standardized methods for verification of the biodegradation process. Based on this literature review, he techniques most commonly employed for this purpose currently include measuring mass loss, examining the surface of plastic fragments by scanning electron microscopy (SEM) and atomic force microscopy (AFM), and using analytical methods such as Fourier transform infrared spectroscopy (FTIR), pyrolysis–gas chromatography–mass spectrometry (Pyr-GC/MS) or high-performance liquid chromatography (HPLC). Each of these methods has its advantages and disadvantages. Nevertheless, currently, there is no universal approach to accurately assess the ability of individual microorganisms to degrade plastics. In this review, we summarize the latest advances in techniques for detecting biodegradation of synthetic polymers and future directions in the development of sustainable strategies for mitigating plastic pollution.

|
Scooped by
?
June 13, 10:39 AM
|
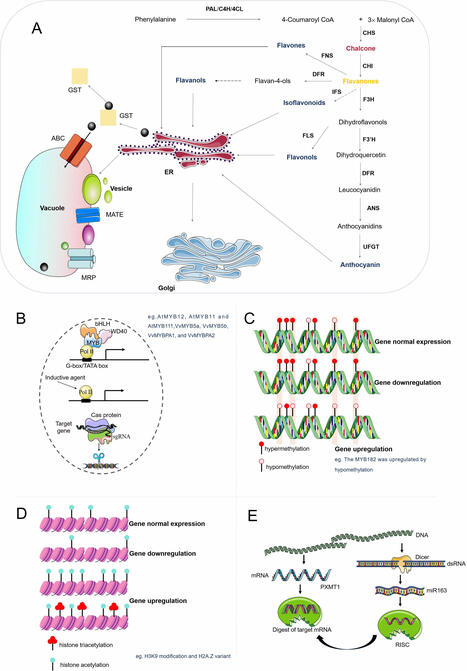
Flavonoids are a diverse class of plant polyphenols with essential roles in development, defense, and environmental adaptation, as well as significant applications in medicine, nutrition, and cosmetics. However, their naturally low abundance in plant tissues poses a major barrier to large-scale utilization. This review provides a comprehensive and forward-looking synthesis of flavonoid biosynthesis, regulation, transport, and yield enhancement strategies. We highlight key advances in understanding transcriptional and epigenetic control of flavonoid pathways, focusing on the roles of MYB, bHLH, and WD40 transcription factors and chromatin modifications. We also examine flavonoid transport mechanisms at cellular and tissue levels, supported by emerging spatial metabolomics data. Distinct from conventional reviews, this review explores how plant cell factories, genome editing, environmental optimization, and artificial intelligence (AI)-driven metabolic engineering can be integrated to boost flavonoid production. By bridging foundational plant science with synthetic biology and digital tools, this review outlines a novel roadmap for sustainable, high-yield flavonoid production with broad relevance to both research and industry.

|
Scooped by
?
June 13, 10:36 AM
|
Plants have evolved complex regulatory mechanisms to cope with environmental challenges. Recent work reveals how distinct root cell types coordinate their specialized genetic responses to form diffusion barriers and adapt to soil stress. In non-compacted soil, water in the soil is absorbed by the roots and transported upwards through the vasculature to support plant hydration and physiological processes. In compacted soil, water stress is exerted on roots as moisture release is reduced from the smaller soil pores. Abscisic acid (ABA) synthesis is activated in the vasculature via unknown genetic mechanisms involving complex hormonal crosstalk with other plant hormones such as ethylene and auxin; the ABA then moves outwards with water flux to activate lignin and suberin deposition in outer tissues. These newly deposited barriers collectively act to prevent root water loss by reducing radial water efflux, preserving internal water status.
|

|
Scooped by
?
Today, 1:05 AM
|
The human body comprises many different microenvironments, each with their own challenges for microorganisms to overcome in order to survive, and possibly cause infection. The human pathogen Streptococcus pneumoniae is notoriously flexible in this regard, and can adapt to a wide range of host niches, including the nasopharynx, lungs, and cerebrospinal fluid. However, the molecular and genetic underpinnings of this ability remain largely obscure. In this work, using infection-mimicking growth conditions we demonstrate that niche adaptation imposes genome-wide changes on multiple levels, including gene essentiality, expression and membrane lipid composition. In general, we show that gene expression and fitness profiling couple orthogonal sets of genes to environmental stimuli. For instance, N-acetylglucosamine (GlcNAc) import (manLMN) and catabolism (nagAB) genes were required for growth on this sugar, but not differentially expressed in its presence, whereas other amino sugar metabolism pathways were upregulated, but not essential. Surprisingly, we found that pneumococci do not necessarily prefer glucose over GlcNAc and that uptake of GlcNAc in absence of subsequent catabolism was toxic. Moreover, we identified a previously overlooked fatty acid saturation regulator, FasR, controlling membrane composition, rendering it important during heat stress. A fundamental understanding of how genes contribute to bacterial niche adaptation, including nutrient availability or temperature fluctuations, is crucial for understanding successful antibiotic therapy and vaccination strategies and the development of novel anti-infectives.

|
Scooped by
?
Today, 12:52 AM
|
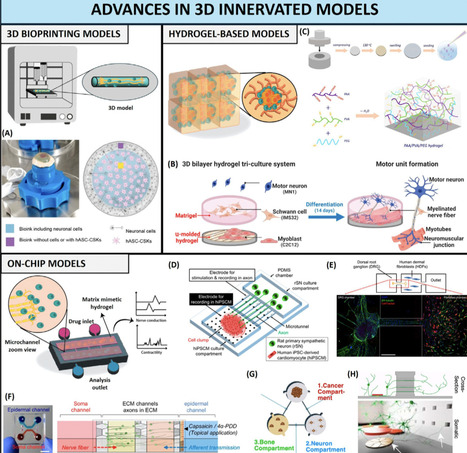
Innervation plays a key role in tissue homeostasis, disease, and repair throughout our lifetimes. Strategies for emulating innervation and its bioinstructive influence on surrounding cells are thus highly desirable to upgrade the organotypic features of human in vitro models. We delve into the latest strategies for generating innervated 3D models that mimic native innervation patterns, and highlight recent advances in bioengineering living platforms for emulating the effect of innervation during tissue regeneration or for recapitulating tumor–nerve interplay. We present an overview of the key design blueprints for generating innervated tissue models, highlight the challenges to be overcome, and discuss the biomedical breakthroughs that can branch from such in vitro platforms.

|
Scooped by
?
Today, 12:37 AM
|
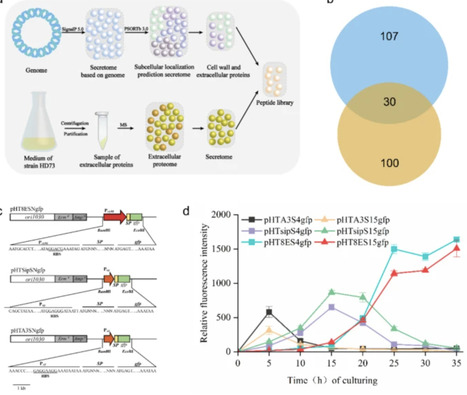
Efficient secretion of heterologous proteins is crucial for applications in industrial and biomedical fields. Selecting appropriate signal peptides and bacterial strains is critical for successful protein expression and export. Bacillus thuringiensis, known for its robust secretion capabilities within the Bacillus genus, shows promise as an ideal host for this purpose. We performed genome-based bioinformatic analysis of B. thuringiensis HD73. A total of 525 proteins were predicted to contain signal peptides, exceeding those in other Bacillus species. The extracellular proteome of B. thuringiensis HD73 was analyzed via LC-MS/MS, identifying 100 secreted proteins. A library of 30 signal peptides was constructed by integrating genome-based predictions with experimental secretome data. Using this library, green fluorescent protein secretory expression systems were developed in the acrystalliferous mutant strain B. thuringiensis HD73−, and the strain carrying signal peptide S17 showed the highest secretion efficiency. Additionally, the top 10 performing signal peptides were used to express and secrete the convenient enzyme cutinase, with the S20 fusion strain exhibiting the highest cutinase activity (3.65 U/mL in the culture supernatant). This study provides the first combined bioinformatic and experimental characterization of the B. thuringiensis secretome. The developed secreted protein expression system and signal peptide library demonstrate effectiveness and offer potential for future heterologous protein secretion in B. thuringiensis.

|
Scooped by
?
Today, 12:06 AM
|
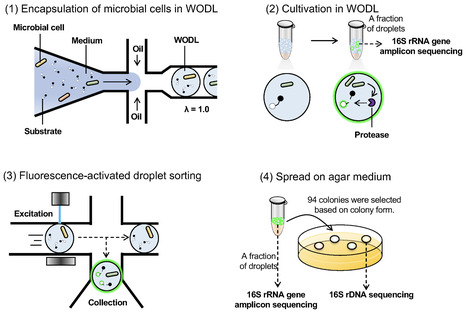
Exploration of diverse microbial sources, particularly environmental bacteria, is needed to identify novel and efficient peptidases/proteases that can be used in a variety of industrial applications. However, conventional function-based screening methods are inefficient and preclude the use of diverse microbial resources. This study illustrates a revolutionary approach to microbial screening using droplet-based microfluidics and fluorescence-activated droplet sorting that targets endopeptidases. Droplet-based microfluidic systems are a powerful tool for culturing microorganisms and for detecting microbial functions inside droplets with ultra-high throughput. However, droplet-based microfluidics for screening the proteolytic activity of environmental bacteria at a large scale remains largely unexplored. Here, we screened approximately 630,000 microorganisms in 6 h and obtained four species with high peptidase activity using droplet-based microfluidics. Furthermore, we isolated an Asp-specific endopeptidase from the isolated bacteria Lysobacter soli and showed that its activity was 2.4-fold higher than that of the related commercially available enzyme. The successful isolation of Asp-specific peptidase with superior activity in a short period of time compared to existing alternatives underscores the efficacy of droplet-based microfluidics for function-based microbial screening.

|
Scooped by
?
June 14, 11:46 PM
|
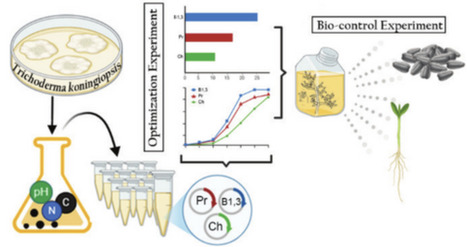
The growing demand for sustainable alternatives to chemical fungicides has driven the development of microbial-based biocontrol strategies. In this study, the native strain Trichoderma koningiopsis LBM116 (Misiones, Argentina) was optimized for the production of mycolytic enzymes (chitinases, β-1,3-glucanases, and proteases) using factorial and response surface experimental designs. Enzyme secretion was increased by more than 250% compared to initial conditions by selecting specific carbon and nitrogen sources and adjusting inoculum and pH parameters. The optimized enzyme formulation improved lettuce seed germination to 86.66% in the presence of the phytopathogen Fusarium sp., under controlled conditions. In seedling trials, it also reduced disease severity and improved growth parameters. These results confirm the dual effect of the enzyme formulation, acting as a biocontrol agent and plant growth promoter. This work highlights the potential of enzyme formulations derived from T. koningiopsis LBM116 as an effective, low-cost, and sustainable alternative for managing phytopathogens in agriculture.

|
Scooped by
?
June 14, 11:15 PM
|
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation of cell function; their engineering, however, is hampered by our limited understanding of allosteric residue networks. Here, we introduce a phage-assisted evolution platform for in vivo optimization of allosteric proteins. It applies opposing selection pressures to enhance activity and switchability of phage-encoded effectors and leverages retron-based recombineering to broadly explore fitness landscapes, covering point mutations, insertions and deletions. Applying our pipeline to the transcription factor AraC yielded optogenetic variants with light-controlled activity spanning ∼1000-fold dynamic range. Long-read sequencing across selection cycles revealed adaptive trajectories and corresponding allosteric interactions. Our work facilitates phage-assisted evolution of allosteric proteins for programmable cellular control.

|
Scooped by
?
June 13, 4:51 PM
|
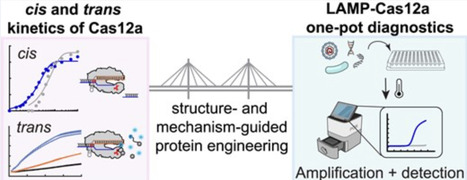
Cas12a trans nuclease activity has been leveraged for nucleic acid detection, often coupled with isothermal amplification to increase sensitivity. However, due to the lack of highly efficient thermostable Cas12a orthologs, use of Cas12a in one-pot combination with high temperature (55–65°C) amplification, such as loop-mediated isothermal amplification (LAMP), has remained challenging. Here, we attempt to address this challenge by comparative study of the thermostable, but poorly trans-active YmeCas12a (from Yellowstone metagenome) with the mesophilic, highly trans-active LbaCas12a (from Lachnospiraceae bacterium ND 2006). Kinetic characterizations identified that poor trans substrate affinity (high Km) is the key limiting factor in YmeCas12a trans activity. We engineered YmeCas12a by structure-guided mutagenesis and fusion to DNA-binding domains to increase its affinity to the trans substrate. The most successful combinatorial variant showed 5–50-fold higher catalytic efficiency with the trans substrate depending on the target site. With the improved variant, we demonstrate efficient nucleic acid detection in combination with LAMP in a single reaction workflow, setting the basis for development of point-of-care tests.

|
Scooped by
?
June 13, 4:33 PM
|
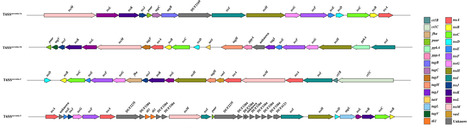
The type VI secretion system (T6SS) is a contact-dependent contractile nanomachine widely distributed among Gram-negative bacteria, essential for interbacterial competition, host interactions and environmental adaptation. The T6SS encoded by Serratia is known to play roles in bacterial competition and contribute to niche adaptation. Yet, the diversity and distribution of T6SS loci and relevant proteins across different Serratia species remain underexplored. In this study, we conducted bioinformatic and comparative genomic analyses of the T6SS to expand our understanding of T6SS in Serratia and their distribution within the genus. A dataset comprising 2,337 Serratia genomes was analysed, identifying 4 distinct T6SS loci classified into 3 main types based on TssB sequence variation, with varying distributions across specific Serratia haplogroups. T6SSserratia-1b subtype was found to be the most prevalent. By integrating homology searches and positional information, we expanded the known database of T6SS proteins in Serratia, identifying a range of effectors likely involved in interbacterial interactions and host adaptation. Additionally, statistical analysis reveals a strong correlation between the presence of T6SS and the diversity of associated genes, including not only known T6SS effectors and immunity proteins but also other co-occurring gene families, enabling the identification of multiple candidate loci potentially involved in bacterial competition and pathogenicity. These findings shed light on the complex distribution, evolutionary dynamics and functional diversity of T6SS and associated proteins in Serratia, advancing our understanding of their role in bacterial competition and environmental adaptation.

|
Scooped by
?
June 13, 3:57 PM
|
Widespread application of bacterial-based cancer therapy is limited because of the need to increase therapeutic bacteria specificity to the tumor to improve treatment safety and efficacy. Here, we harness the altered tumor metabolism and specifically elevated kynurenine accumulation to target engineered bacteria to the cancer site. We cloned and leveraged kynurenine-responsive transcriptional regulator (KynR) with its cognate promoter in Escherichia coli. Optimizing KynR expression coupled with overexpressing kynurenine transporter and amplifying the response through plasmid copy number–based signal amplification enabled the response to kynurenine at the low micromolar levels. Knocking out genes essential for cell wall synthesis and supplying these genes via kynurenine-controlled circuits allowed tuning Salmonella enterica growth in response to kynurenine. Our kynurenine-controlled S. enterica (hereafter named AD95+) showed superior tumor specificity in breast and ovarian cancer murine models compared to S. enterica VNP20009, one of the best characterized tumor-specific strains. Last, AD95+ showed anticancer properties compared to vehicle controls, demonstrating the potential as an anticancer therapeutic.

|
Scooped by
?
June 13, 11:19 AM
|
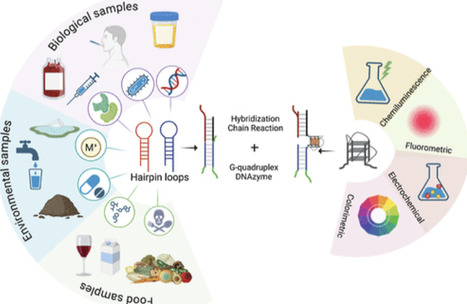
The hybridization chain reaction (HCR) is an amplification method recognized for detecting analytes present in trace quantities owing to its high specificity, sensitivity, and straightforward approach. Simultaneously, G-quadruplex DNAzymes augment HCR-based biosensors, serving as transducers due to their elevated catalytic activity and nonenzymatic methodology. The current review aims to provide readers with a critical overview of significant aspects of biosensors that utilize HCR for analyte detection amplification and G-quadruplex as transducers, focusing on the latest activities of global researchers. A bibliometric analysis was conducted to identify key research topics in HCR and G-quadruplex-based sensors. Further, the review aims to elucidate the advantages and disadvantages of each strategy employed for detection using this sort of sensor. Specific sections have been included to critically evaluate the potential of these biosensors across many scientific domains. In conclusion, the limitations of the existing sensor that restrict its applicability in real-time scenarios have been identified for subsequent enhancement. Plausible strategies that could enhance these sensors and emerging scientific fields that could benefit from HCR and G-quadruplex DNAzyme-based sensors have been elucidated.

|
Scooped by
?
June 13, 10:41 AM
|
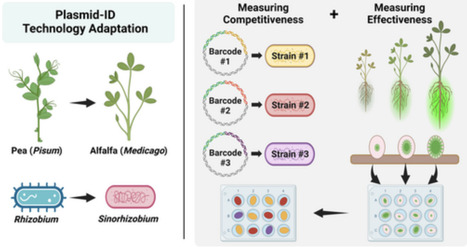
Maximizing the nitrogen fixation occurring in rhizobia-legume associations represents an opportunity to sustainably reduce nitrogen fertilizer inputs in agriculture. High-throughput measurement of symbiotic traits has the potential to accelerate the identification of elite rhizobium/legume associations and enable novel research approaches. Plasmid-ID technology, recently deployed in Rhizobium leguminosarum, facilitates the concurrent assessment of rhizobium nitrogen-fixing effectiveness and competitiveness for root nodulation. This study adapts Plasmid-ID technology to function in Sinorhizobium species that are central models for studying rhizobium-legume associations and form economically important symbioses with alfalfa. New Sino-Plasmid-IDs were developed and tested for stability and their ability to measure competitiveness for root nodulation and nitrogen-fixing effectiveness. Rhizobial competitiveness is measured by identifying strain-specific nucleotide barcodes using next-generation sequencing, whereas effectiveness is measured by GFP fluorescence driven by the synthetic nifH promoter. Sino-Plasmid-IDs allow researchers to efficiently study competitiveness and effectiveness in a multitude of Sinorhizobium strains simultaneously.

|
Scooped by
?
June 13, 10:37 AM
|
One of the main goals in synthetic biology is to produce value-added compounds from available precursors using enzymatic approaches. The construction of biosynthetic pathways for synthesizing target molecules plays a crucial role in this process. However, it is challenging and time-consuming for researchers to design efficient pathways manually. In recent decades, pathway design has advanced through data- and algorithm-driven approaches. In this article, we review key computational tools involved in biosynthetic pathway design, covering: 1) Biological Big-Data including compounds, reactions/pathways and enzymes. 2) Retrosynthesis methods leveraging multi-dimensional biosynthesis data to predict potential pathways for target compounds synthesis. 3) Enzyme engineering relying on data mining to identify/de novo design enzymes with desired functions. Integrating these three key components can significantly enhance the efficiency and accuracy of biosynthetic pathway design in synthetic biology.
|
 Your new post is loading...
Your new post is loading...



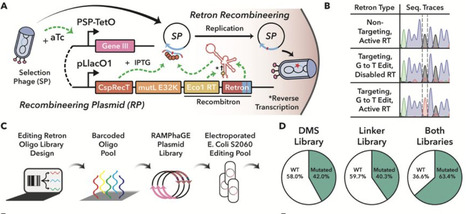










3st, directed evolution, existing phage-assisted evolution systems focus on optimizing a selected activity under more or less static conditions, rendering them ill-suited for evolving proteins that must switch between distinct functional states in response a defined input. POGO-PANCE (Protein-switch On/Off Gene Optimization using Phage-Assisted Non-Continuous Evolution): POGO-PANCE cyclically “bounces” evolving proteins between alternating selection steps corresponding to the presence and absence of a defined input signal. mutagenesis plasmids (MPs) increase point mutation rates by perturbing DNA replication and repair (22). However, they introduce strong mutational biases and cannot generate insertions or deletions. developed RAMPhaGE (Recombitron-Assisted Multiplexed Phage Gene Evolution), a retron-based in vivo diversification method capable of introducing point mutations and InDels at tunable frequencies. effector activity promotes expression of M13 gene III (gIII), encoding an essential phage coat protein (pIII), from an accessory plasmid (AP) in trans, thereby enabling phage propagation (15). In contrast, during negative selection, the same regulatory circuit is inverted such that effector protein activity drives expression of a dominant-negative gIII variant (gIII-Neg), which is incorporated into assembling phage particles and blocks their release from the host (23).
To turn retrons into a targeted in vivo diversification strategy of M13 phage carried transgenes, we used a Recombitron: a fully genetically encoded operon combining a retron, single-stranded annealing protein (CspRecT), and mutL, a dominant-negative variant of the E. coli mismatch repair suppression protein mutS