Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 5:46 PM
|
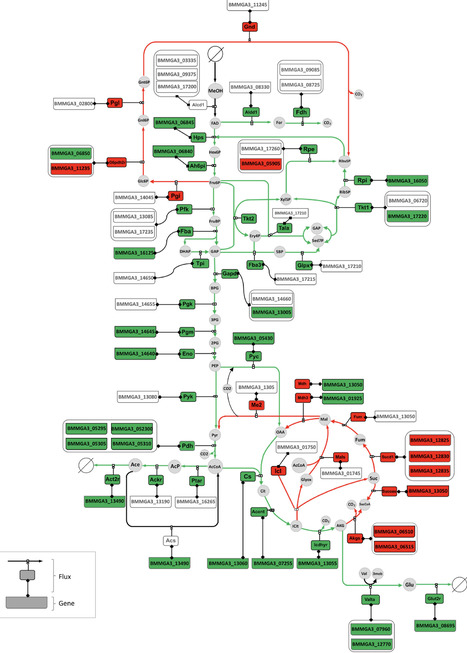
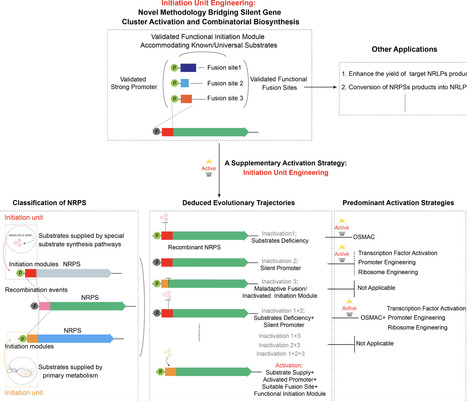
Nonribosomal peptide synthetases (NRPS) represent a valuable yet underexplored resource for producing bioactive natural products. However, most NRPSs remain silenced potentially due to factors such as dysfunction of the initiation unit. The starter condensation (Cs) domain of the initiation unit catalyzes the lipoinitiation of nonribosomal peptides via the incorporation of an N-terminal fatty acyl chain. The concept of initiation unit engineering introduced herein encompasses the replacement of the native initiation unit of NRPSs with a foreign and well-characterized Cs domain-containing initiation unit to activate the NRPS and optimize its expression. This strategy was employed herein to successfully access three of the six previously silent NRPS pathways in Mycetohabitans rhizoxinica HKI 454, a bacterium of the class β-proteobacteria, resulting in the identification of three classes of lipopeptides. This strategy was then extended to access two NRPS pathways in Pseudomonas syringae (γ-proteobacteria) and obtain novel lipopeptides, thereby establishing a feasible complement to existing genome mining strategies for natural product discovery. Furthermore, change of the initiation regions of biosynthetic pathways of nonlipidated chitinimide (β-proteobacteria) and pseudotetraivprolide (γ-proteobacteria) with heterologous Cs-containing initiation units enabled the successful incorporation of fatty acyl chains into the N-terminus of both peptide backbones, launching a workable approach to create artificial lipopeptides. Overall, this study provides a practical strategy for the rational recovery of silent BGCs and introduction of fatty acyl chains into nonribosomal peptides, at least in Proteobacteria, thereby enriching genome mining and combinatorial biosynthesis approaches for accessing the underexplored biosynthetic potential of NRPSs from various bacteria.

|
Scooped by
mhryu@live.com
Today, 5:33 PM
|
Synthetic microbial communities (SynCom) are microbial consortia with defined taxonomic and functional traits, so that the combination elicits a predictable response under defined conditions. SynComs are artificially designed to enable inter-species metabolic interactions, metabolic division of labor, and ecological interactions that can elicit phenotypes like colonization stability and environmental adaptation. As an applied tool, SynComs have been deployed in diverse contexts, including agriculture, industry, and environmental ecology. This systematic review explores the processes used to construct SynComs, the mechanisms of metabolic interaction between members, and a review of the different ways that SynComs have been applied. We also explore the challenges for SynCom development and application, and future research directions that could overcome these challenges. SynComs are a powerful tool in our arsenal of applied technologies, but research and application are still nascent. While advances have been made, more research is needed to ensure SynCom technologies do not threaten global ecological security. SynCom technology represents a versatile platform for the controlled manipulation of microbial systems, enabling targeted modification of ecological and physiological processes. This emerging field marks a transition from descriptive biology toward a predictive and engineering-driven framework for understanding and shaping living systems.

|
Scooped by
mhryu@live.com
Today, 5:21 PM
|
Macrophages show great potential for application in cellular immunotherapy, but are limited by immune checkpoints (ICBs). Immune checkpoint inhibitors (ICIs) can effectively block immune escape pathways and alleviate immune suppression in the tumor microenvironment (TME). Here, we constructed hypoxia response bacteria (HRB) that specifically expressed CD47 antibody (aCD47) under hypoxic conditions in the TME, enabling the in situ synthesis of ICIs at the tumor site. In addition, this study further prepared responsive liposomes for encapsulating the STING agonist cGAMP and modified them by covalent attachment to the HRB surface to form the composite material HRB@LC. This composite system can synergistically block CD47-SIRPα-mediated immune escape and activate the STING signal pathway, thereby enhancing systemic antitumor immune responses and significantly improving the efficacy of immunotherapy.

|
Scooped by
mhryu@live.com
Today, 5:13 PM
|
In Bacillus species, the signal peptide (SP) efficiently guiding the protein secretion is crucial for production, yet there is still a lack of reliable computational tools to accurately predict its efficiency. Therefore, we developed SecEff-Pred, a novel web server that leverages an ESM-2-based predictor. Enhanced by an innovative data simulation strategy and a multitask learning framework, SecEff-Pred accurately predicted the secretion efficiency of signal peptides in Bacillus subtilis. The server demonstrated exceptional performance, achieving prediction accuracies of 85.59% for α-amylase, 81.58% for alkaline xylanase, and 74.68% for cutinase. SecEff-Pred was further validated using phospholipase D (PLD) as a reporter protein, demonstrating high prediction accuracy for signal peptide efficiency (overall 72%), with an accuracy of 80% for “efficient” SPs (corresponding to a maximum PLD activity of 929 U/mL) and 62.50% for “inefficient” SPs. These results confirm the SecEff-Pred is a powerful tool for guiding protein secretion in B. subtilis.

|
Scooped by
mhryu@live.com
Today, 4:54 PM
|
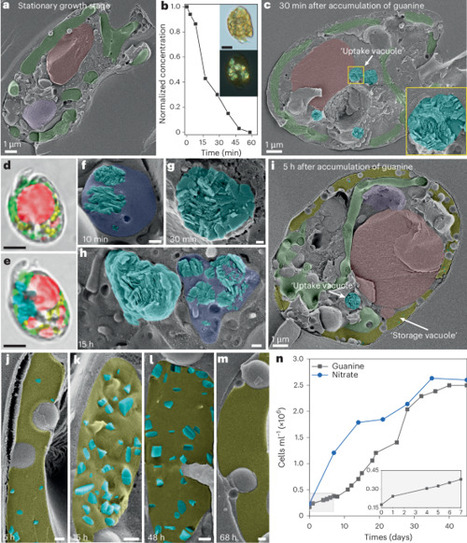
Highly reflective biogenic crystals such as guanine have potential as biocompatible alternatives to toxic inorganic optical materials. However, controlling the structural and optical properties of these sparingly soluble crystals in vitro is challenging. Engineered microbial cells have been used widely to generate high-value metabolites, but the biosynthesis of functional crystalline materials has not been achieved. Here we harness microalgae for the biosynthesis of difficult-to-crystallize molecular materials. We show that dinoflagellates can rapidly accumulate many nitrogen heterocycles from aqueous solutions into nitrogen-storage crystals, revealing a general mechanism for their metabolism of dissolved organic nitrogen. We manipulate this innate crystallization behavior to generate crystals with tailored morphologies and optical properties, including birefringent xanthine spherulites—a biogenic analog of TiO2 nanoparticles. Our results show how microalgae may be exploited as cellular factories for producing molecular crystals from aqueous solutions, under ambient conditions, harnessing the intrinsic control mechanisms of crystal-forming cells, with possible further applications in the crystallization of pharmaceuticals and bioremediation of toxicants. Crystalline materials for optical applications are synthesized in living dinoflagellates.

|
Scooped by
mhryu@live.com
Today, 4:31 PM
|
Nearly two years after Google DeepMind released an updated AlphaFold3 geared at drug discovery, its biopharmaceuticals spin-off, Isomorphic Labs, announced an even more powerful artificial-intelligence model — and they’re keeping it all to themselves. Isomorphic Labs, based in London, touted the capacities of its ‘drug-discovery engine’ — which it calls IsoDDE — in a 27-page technical report, released on 10 February. Achievements, including precise predictions of how proteins interact with potential drugs and antibody structures, have impressed scientists working in the field. An open-source model called Boltz-2, developed by scientists at MIT and released last year, could predict the strength to which potential drugs glom onto proteins, or binding affinity.

|
Scooped by
mhryu@live.com
Today, 4:22 PM
|
Synthetic biology aims to achieve predictable, programmable control over living systems by designing and engineering biological components and functions. Over the past 25 years, the field has advanced from foundational molecular tools to increasingly complex systems-level architectures. A new inflection point has emerged with the integration of generative artificial intelligence (AI), catalyzing a fundamental shift in how biological design is conceived and executed. Generative AI now enables the data-driven creation of novel designs with predictable functionality and context-aware precision. Here, we examine the convergence of synthetic biology and generative AI, highlighting key innovations at this emerging frontier of deep generative design across biological parts and systems. We discuss how design frameworks have evolved and outline the opportunities and challenges that lie ahead, spanning biomolecular elements, genetic circuits, and genomes. Finally, we propose a roadmap for how generative AI can unlock a new era of predictable, programmable synthetic biological systems.

|
Scooped by
mhryu@live.com
Today, 1:21 PM
|
The preference in synonymous codon usage—the so-called codon usage bias (CUB)—is governed by several factors such as the host organism, context and function of the gene, and the position of the codon within the gene itself. We demonstrated that this mapping can be learned from the host’s genome using language models and subsequently applied for codon optimization of heterologous proteins expressed by the host. This pipeline called Pichia–Codon language model (Pichia-CLM) was applied to the industrial host organism, Komagataella phaffii. With this approach, production of heterologous proteins was enhanced up to threefold compared to their native sequences. Furthermore, Pichia-CLM consistently yielded constructs with enhanced productivity for proteins of varied complexity, compared to commercially available tools. Finally, we showed that Pichia-CLM generates sequences resembling the properties of codon usage found in the host’s intrinsic host cell proteins and learned features such as avoiding negative cis-regulatory and repeat elements based on patterns in the genome data. These results show the potential of language models to unbiasedly learn patterns and design robust sequences for improved protein production.

|
Scooped by
mhryu@live.com
February 18, 11:58 PM
|
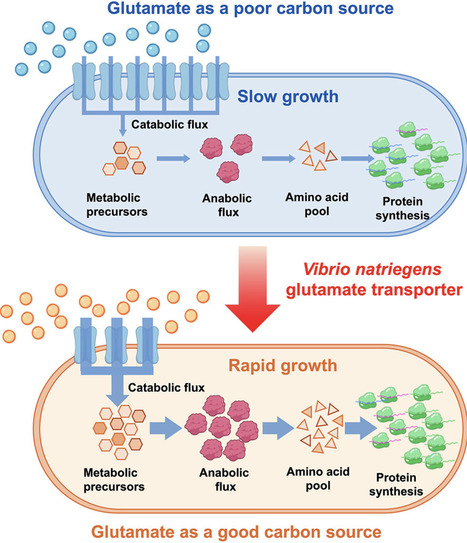
Bacterial growth modulation is crucial in microbial synthetic biology. In this study, we found that glutamate is an extremely poor carbon source for E. coli and Bacillus subtilis. The slow growth on glutamate can be effectively overcome by the heterologous expression of glutamate transporters from Vibrio natriegens. Our results revealed that cross-species substrate transporters could be employed to shift bacterial cellular resource allocation, offering a potential genetic strategy for modulating microbial biomass growth.

|
Scooped by
mhryu@live.com
February 18, 11:47 PM
|
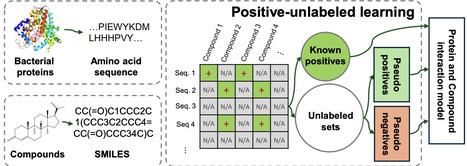
Prediction of Compound-Protein Interactions (CPI) in bacteria is crucial to advance various pharmaceutical and chemical engineering fields, including biocatalysis, drug discovery, and industrial processing. However, current CPI models cannot be applied for bacterial CPI prediction due to the lack of curated negative interaction samples. We propose a novel Positive-Unlabeled (PU) learning framework, named BIN-PU, to address this limitation. BIN-PU generates pseudo positive and negative labels from known positive interaction data, enabling effective training of deep learning models for CPI prediction. We also propose a weighted positive loss function that weights to truly positive samples. We have validated BIN-PU coupled with multiple CPI backbone models, comparing the performance with the existing PU models using bacterial cytochrome P450 (CYP) data. Extensive experiments demonstrate the superiority of BIN-PU over the benchmark models in predicting CPIs with only truly positive samples. Furthermore, we have validated BIN-PU on additional bacterial proteins obtained from literature review, human CYP datasets, and uncurated data for its reproducibility. We have also validated the CPI prediction for the uncurated CYP data with biological and biophysical experiments. BIN-PU represents a significant advancement in CPI prediction for bacterial proteins, opening new possibilities for improving predictive models in related biological interaction tasks.

|
Scooped by
mhryu@live.com
February 18, 5:00 PM
|
Intrinsically disordered proteins and regions (collectively IDRs) are found across all kingdoms of life and have critical roles in virtually every eukaryotic cellular process. IDRs exist in a broad ensemble of structurally distinct conformations. This structural plasticity facilitates diverse molecular recognition and function. Here we combine advances in physics-based force fields with the power of multi-modal generative deep learning to develop STARLING, a framework for rapid generation of accurate IDR ensembles and ensemble-aware representations from sequence. STARLING supports environmental conditioning across ionic strengths and demonstrates proof of concept for the interpolative ability of generative models beyond their training domain. Moreover, we enable ensemble refinement under experimental constraints using a Bayesian maximum-entropy reweighting scheme. Beyond ensemble characterization, STARLING sequence representations can be used in multiple ways. We showcase two examples: first, STARLING lets us perform ensemble-based search for ‘biophysical look-alikes’. Second, we demonstrate how these latent representations can be used to accelerate ensemble-first sequence design from weeks or hours per candidate to seconds, enabling library-scale designs. Together, STARLING dramatically lowers the barrier to the computational interrogation of IDR function through the lens of emergent biophysical properties, complementing bioinformatic protein sequence analysis. We evaluate the accuracy of STARLING against extant experimental data and offer a series of vignettes illustrating how STARLING can enable rapid hypothesis generation for IDR function and aid the interpretation of experimental data. The deep learning model STARLING can generate accurate ensembles of intrinsically disordered regions of proteins using only protein sequence as input.

|
Scooped by
mhryu@live.com
February 18, 4:48 PM
|
Biological functions depend on the spatiotemporal distribution of proteins within cells. Key cellular activities such as signal transduction, metabolism, cell cycle and cell death are driven by the interactions of proteins that are localized in multiple cellular compartments. Such multilocalization can even allow protein with identical sequences to display multifunctionality, a phenomenon known as moonlighting. Despite its biological importance, the relationship between protein localization and function remains underexplored. In this Review, we discuss the known mechanisms of protein localization (including RNA transport, role of proteoforms and molecular interactions) and how subcellular localization controls protein function. Proper regulation of protein localization is crucial for specialized cell and tissue functions, including cell differentiation, polarization and the epithelial–mesenchymal transition. Protein mislocalization can also have important roles in pathological processes, such as in cancer, neurodegeneration and autoimmunity. We end with a discussion of current technological and conceptual challenges in the field of subcellular proteomics and spatial biology. Addressing these challenges will allow us to link the dynamic nature of protein localization and function across biological scales and contexts, with great impact on fundamental cell biology and clinical applications. Proteins can localize to multiple cellular compartments and some exhibit distinct functions depending on their location. This Review discusses the mechanisms of protein localization, the control of specialized protein functions through subcellular localization, and how mislocalization is involved in cancer, neurodegeneration and autoimmunity.

|
Scooped by
mhryu@live.com
February 18, 1:52 PM
|
An outstanding challenge in molecular biology is the production of a complete and accurate set of protein interactions for a given organism. Todor et al, introduce a clever strategy to efficiently screen all protein interactions in Mycoplasma genitalium using AlphaFold3 (Todor et al, 2026).
|

|
Scooped by
mhryu@live.com
Today, 5:39 PM
|
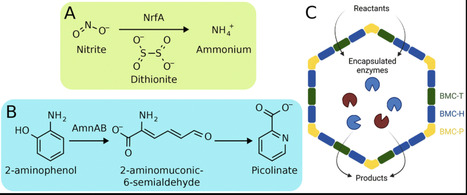
Bacterial microcompartments (BMCs) are protein-based organelles found in diverse bacteria that encapsulate a sequence of enzymes to accelerate specific metabolic pathways and limit toxicity by confining intermediate products. To facilitate catalysis in BMCs, reactants and products must transit across the protein shells, which are made up of multimeric protein tiles. Quantifying the permeability for transport of small molecules across the shell is a key engineering consideration to design novel catalytic microcompartments. We examine the permeability of reactants and products for two reaction pathways, the degradation of 2-aminophenol by AmnAB and the reduction of nitrite to ammonium by NrfA, through a computational lens to quantify permeability for these metabolites at the nanoscale. From Hamiltonian replica exchange umbrella sampling simulations, we determine that the energetic barriers to permeation for the relevant metabolites along both pathways are small, with permeability coefficients in the range of 0.1–4 cm s−1. The high permeabilities calculated for each metabolite through the inhomogeneous solubility diffusion model are corroborated by enzyme activity measurements in vitro that point to enzymatic activity within the shell system. We expand on these findings by quantifying the scale of transport and catalysis that can be supported by in vitro BMC systems. The results highlight the importance of substrate permeability across BMCs within a biological context, and represent steps toward generating synthetic shells or bioengineering novel nanoreactors.

|
Scooped by
mhryu@live.com
Today, 5:29 PM
|
Cancer immunotherapy is increasingly moving toward personalized, precision-based strategies, with cancer vaccines emerging as a promising approach to reshape treatment. However, despite their potential, current tumor vaccines often yield limited clinical responses and subpar immunogenicity, underscoring the urgent need for innovative delivery systems to enhance immune activation. Bacterial outer membrane vesicles (OMV), which possess natural immunomodulatory properties and impressive engineering flexibility, have attracted attention as versatile platforms for vaccine development and bioengineering applications. This review thoroughly summarizes recent advances in using OMVs to enhance the effectiveness of cancer vaccines. First, we explain the key biological features of OMVs that support their immunotherapeutic potential. Next, we carefully analyze the primary mechanisms by which OMVs enhance immune responses, as well as cutting-edge engineering strategies to improve their safety, immunogenicity, and specificity. Additionally, we discuss the significant challenges that hinder the clinical use of OMV-based cancer vaccines and provide a comprehensive review of current progress and future outlooks. Looking forward, combining artificial intelligence, tumor microenvironment profiling, and neoantigen discovery is expected to drive the development of next-generation, personalized OMV-based immunotherapies. Overall, OMVs stand out as a transformative platform capable of overcoming major obstacles in cancer vaccine development and pushing forward future cancer immunotherapy.

|
Scooped by
mhryu@live.com
Today, 5:16 PM
|
Phage therapy is an attractive countermeasure to multidrug-resistant pathogens, but clinical deployment is limited by the narrow and poorly predictable host range of most phages. Public repositories now house tens of thousands of phage genomes, yet there is no systematic route for turning this sequence space into designer phages with defined receptor specificities. Here we present a scalable, data-driven framework that converts receptor-binding protein (RBP) diversity into a modular toolkit for programmable phage engineering. Using Klebsiella pneumoniae as a model, we mined 280 non-redundant Przondovirus RBP sequences and resolved them into >50 discrete clusters. Functional screening of the Prz_RBPs from 41 previously uncharacterized Przondovirus phages expanded the number of experimentally validated capsular locus (KL) targets from 14 to 32 in Przondovirus. Each cluster was primarily associated with a dominant KL type, enabling construction of a genotype-to-phenotype map that accurately predicts receptor tropism. The identification of conserved anchor motifs enabled combinatorial pairing of Prz_RBP1 and Prz_RBP2 as plug-and-play modules for programmable phage tropism. Supplying exogenous Prz_RBP2 variants that assemble with Prz_RBP1 further and predictably expands host range, providing broad and tunable coverage. This framework transforms raw genomic diversity into customizable antibacterial agents and offers a general blueprint for precision phage therapy.

|
Scooped by
mhryu@live.com
Today, 5:07 PM
|
Preozonation is widely used to enhance the effectiveness of coagulation and filtration in algae-laden water treatment, but cyanobacterial cell rupture and the subsequent release of intracellular organic matter and cyanotoxins can increase treatment burdens and pose health risks. In natural waters, cyanobacteria are often surrounded by symbiotic bacteria, whose influence on ozonation performance and underlying mechanisms remains unclear. Herein, we found that axenic filamentous cyanobacteria (Leptolyngbya sp.) exhibited strong resistance to ozonation (0.3 mg L–1, 20 min), whereas the presence of surface-associated bacteria markedly increased the cell rupture rate from 12 ± 6% to 76 ± 2%. Removal of loosely bound extracellular polymeric substances (LB-EPS) significantly reduced ozonation resistance in axenic cyanobacteria but unexpectedly enhanced that of xenic cultures. By integrating reactive oxygen species identification, extracellular metabolomics, and metabolic reconstruction, we demonstrate that surface-colonizing bacteria degrade the algal LB-EPS envelope, releasing metabolites that facilitate hydroxyl radical formation during ozonation, thereby intensifying cell rupture. Our results highlight surface-associated bacteria as a critical yet overlooked factor shaping cyanobacterial responses to preozonation, underscoring the need to re-evaluate ozone application strategies in bloom-impacted waters to minimize cell rupture and byproduct formation.

|
Scooped by
mhryu@live.com
Today, 4:47 PM
|
Small-molecule metabolites are key pharmaceutical resources embedded in complex organismal metabolomes. Scalable microbial production depends on metabolic activation capacity, which in turn requires efficient genetic variation. Structural variants (SVs), key drivers of phenotypic diversity, are pivotal for organism evolution, yet their highly efficient induction remains challenging. While DNA double-strand breaks (DSBs) facilitate SVs formation, existing mutagenesis technologies struggle to balance high DSB efficiency with cellular preservation, particularly in microbial strain improvement for metabolite production. Conventional irradiation methods suffer from low SVs induction rates, making strain enhancement a lengthy and labor-intensive process. Here, we systematically compare six irradiation technologies in Streptomyces lividans 1326 and identify high-energy pulsed electron beams (HEPE) as an approach which effectively induces strong DSBs while preserving cellular integrity. This results in extensive SVs that reshape genome sequences and 3D chromatin structure, leading to activation of secondary metabolite production. By integrating HEPE with high-throughput metabolomics (HEPE-HiTMS), we discover two secondary metabolites with unusual C-N linkage, respectively. Applied across various microorganisms, HEPE enables record-high clavulanic acid and microcin J25 production, and markedly increases lovastatin yields. With its ability to induce SVs with minimal cytotoxicity, HEPE represents a powerful tool for cryptic metabolite discovery and industrial strain development. The majority of microbial metabolites remain cryptic or minimally expressed under laboratory conditions. Here authors show that high-energy pulsed electron beams (HEPE) can be used to induce extensive structural variation, activating secondary metabolite biosynthesis.

|
Scooped by
mhryu@live.com
Today, 4:25 PM
|
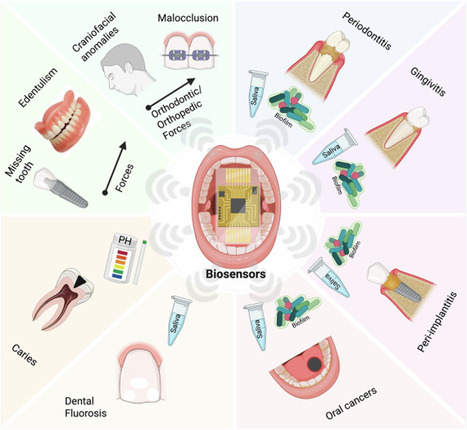
Oral and dental health is an important indicator and determinant of an individual’s overall well-being. Untreated oral diseases can lead to severe systemic complications. Monitoring the oral environment and identifying biochemical and physiological patterns associated with disease states, such as periodontitis, gingivitis, caries, and oral cancers, is essential for early diagnosis and effective intervention. This review evaluates the current clinical needs in biochemical and physiological monitoring for oral healthcare and state-of-the-art biosensors capable of continuous analyte measurement. We surveyed the relevant biomarkers for common oral and dental diseases in patients compared to healthy controls. The design and performance of recent biosensing devices for these target analytes are reviewed and evaluated. For biochemical sensing, we found intraoral biosensors for high-abundance small molecules, such as ions and metabolites, have advanced significantly in recent years. However, robust sensing technologies for low-abundance analytes, including cytokines and other inflammatory biomarkers, remain limited and require further development in sensing mechanisms, bio-interfaces, and device integration. For physiological sensing, particularly the measurement of forces in tooth movement, recent developments in force sensor technologies have substantially improved measurement accuracy over traditional techniques. Despite these advancements, current platforms still face limitations in achieving long-term, real-time monitoring of mechanical conditions within the oral cavity due to challenges related to biocompatibility and device miniaturization. In conclusion, while notable progress has been made in biosensing for oral applications, continued research in device integration with clinical practices is essential to realize robust and clinically deployable biosensor systems that can advance precision oral healthcare.

|
Scooped by
mhryu@live.com
Today, 4:12 PM
|
Rooted in place, plants must continuously respond and adapt to their ever-changing environment to survive, especially as climate change intensifies. Calcium ions (Ca²⁺) play a central role in plant responses to both biotic and abiotic challenges. Ca²⁺ signaling involves the coordinated action of channels and transporters that generate specific “Ca²⁺ codes,” along with Ca²⁺-binding proteins that act as sensors to decode them. Studies over the past several decades have explored the molecular components that form the toolkit, pathways, and networks for the coding and decoding of Ca²⁺ signals in plants. This review focuses on the emerging mechanisms of calcium signaling in plants, beginning with an overview of the universal conceptual framework that governs the coding and decoding of Ca²⁺ signals, followed by examples of pathways in plant growth and reproduction, responses to abiotic stress and microbes, and systemic signaling in plants.

|
Scooped by
mhryu@live.com
Today, 12:17 AM
|
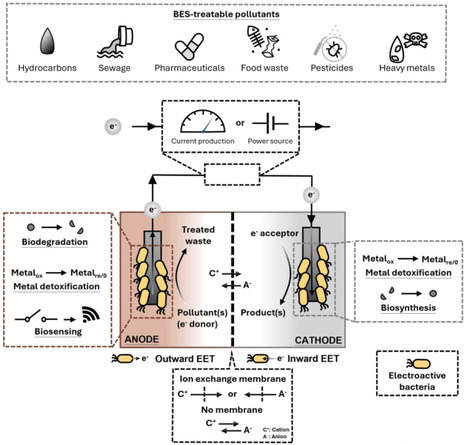
Electroactive bacteria (EAB) can exchange electrons with conductive materials as part of their metabolic activity, enabling the development of diverse bioelectrochemical systems (BESs). These systems can be used for sustainable power generation, pollutant biosensing, and pollutant bioremediation. Here, we first discuss recent studies that expand the bioremediation and biosensing capabilities of lab-scale BESs by using native and engineered EAB and microbial consortia. Then, we review innovative strategies implemented by large-scale pilot studies and startup companies to scale up BESs for low-cost bioremediation. This review summarizes the complementary insights gained from research done at these different scales and discusses where this knowledge can take us.

|
Scooped by
mhryu@live.com
February 18, 11:53 PM
|
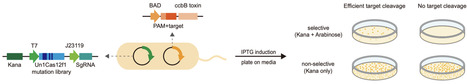
The compact CRISPR-Cas12f system is promising for AAV-delivered gene therapy, but its application has been constrained by restrictive PAM recognition (e.g., TTTR) and suboptimal editing efficiency. Through bacterial library screening and mammalian cell validation, we engineer evoCas12f, an optimized variant incorporating five key mutations, that dramatically expands PAM recognition to NTNR/NYTR. This advancement reduces median distance between two neighbouring PAM sites to 2 nucleotides in the human genome. It also demonstrates 1.4-fold enhanced activity at TTTR sites compared to wild-type Un1Cas12f1, achieving up to 91% editing efficiency. Remarkably, evoCas12f enables efficient generation of homozygous mutations in F0 generation mice, even at non-canonical PAM sites. We further adapt this system for robust transcriptional activation and precise base editing with a well-defined editing window. As a compact yet highly efficient platform, evoCas12f represents a significant advance in CRISPR technology, enabling multiplexed editing for high-resolution targeting applications and expanding possibilities for therapeutic genome engineering. Compact Un1Cas12f1 enzyme suits AAV delivery but is limited by narrow PAMs and modest activity. Here, authors engineer evoCas12f to recognize broader NTNR/NYTR PAMs and boost editing efficiency up to 91%, enabling efficient multiplex editing, base editing, and gene activation in cells and mice.

|
Scooped by
mhryu@live.com
February 18, 11:40 PM
|
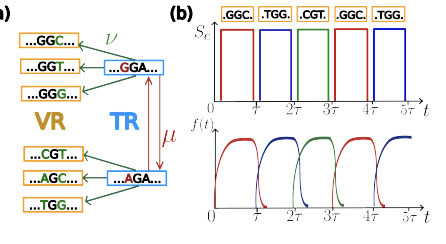
Diversity-Generating Retroelements (DGRs) create rapid, targeted variation within specific genomic regions in phages and bacteria. They operate through stochastic retro-transcription of a template region (TR) into a variable region (VR), which typically encodes ligand-binding proteins. Despite their prevalence, the evolutionary conditions that maintain such hypermutating systems remain unclear. Here we introduce a two-timescale framework separating fast VR diversification from slow TR evolution, allowing the dynamics of DGR-controlled loci to be analytically understood from the TR design point of view. We quantity the fitness gain provided by the diversification mechanism of DGR in the presence of environmental switching with respect to standard mutagenesis. Our framework accounts for observed patterns of DGR activity in human-gut Bacteroides and clarifies when constitutive DGR activation is evolutionarily favored.

|
Scooped by
mhryu@live.com
February 18, 4:54 PM
|
Pseudomonas species display exceptional metabolic versatility that underpins their ecological success and broad relevance for synthetic biology, environmental microbiology, bioremediation, and bioproduction. A central contributor to this versatility is the ped gene cluster, which encodes pyrroloquinoline quinone (PQQ)-dependent dehydrogenases that catalyze the oxidation of a wide range of alcohols and aldehydes. These enzymes support both assimilation and detoxification processes with high catalytic efficiency. This review compiles current knowledge on genetic organization, enzymatic functions, and multi-level regulation of the ped cluster, with a focus on Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. The roles of regulatory components [e.g., the iron (Fe2+)-dependent YiaY dehydrogenase and the hybrid PP_2683 histidine kinase] are examined for their capacity to respond to short-chain alcohols through a complex signal transduction network. Additional genetic elements, including pedF and pedG, along with poorly characterized open reading frames (e.g., pedD, PP_2666, and PP_2678), which support enzymatic maturation, electron flow, and modulation of surface-associated behaviors are likewise considered. Comparative analysis across the Pseudomonas genus showed that ped-like clusters are conserved but display substantial differences in gene content and arrangement, suggesting adaptations to specific ecological contexts. We evaluate these elements in detail to define a reference framework for future mechanistic studies. By bringing together functional and regulatory features of the cluster, our article provides a basis for exploiting the Ped system as a modular platform in applied microbiology. This integrated view aims to guide ongoing and future fundamental and applied research on alcohol oxidation in gram-negative bacteria.

|
Scooped by
mhryu@live.com
February 18, 3:06 PM
|
Anabaena, a nitrogen-fixing cyanobacterium from the Nostocaceae family, is a promising chassis for sustained space exploration. Using NEKO, this study surveys 2000 publications on Anabaena and summarizes the research, including systems biology, modeling, and potential space applications. Future research should examine its metabolism in space environments, such as microgravity and radiation, and develop bioreactor designs and genetic tools for reliable, self-regulating biomanufacturing systems in these environments.
|
 Your new post is loading...
Your new post is loading...