Your new post is loading...

|
Scooped by
mhryu@live.com
January 23, 5:29 PM
|
To counter challenges from bacteriophages (phages), bacteria use defense mechanisms that can reside on mobile genetic elements or within chromosomes. These immune systems are easily gained and lost, allowing adaptation to threats. However, the mechanism of mobilization of chromosomally encoded defense genes remains poorly understood. Here, we show that phage- and phage-inducible chromosomal island (PICI)–mediated lateral transduction (LT), a highly efficient horizontal gene transfer HGT mechanism, facilitates the transfer of these defense genes between bacteria. Using several bacterial models, we demonstrate that defense systems are often positioned near phage or PICI attachment sites, allowing them to exploit LT for their mobility. In addition, LT diversifies defense genes carried by prophages and PICIs, driving immune system evolution and turnover. These processes provide phage resistance to new bacterial hosts and profoundly affect population genomics. Our findings reveal LT as a crucial mechanism shaping bacterial evolution and influencing the trajectory of pathogenic clones in nature.

|
Scooped by
mhryu@live.com
January 23, 5:24 PM
|
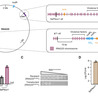
Terpenoid-based biofuels represent a sustainable alternative to fossil fuels with superior energy densities and combustion properties. However, achieving industrial-scale production requires overcoming multiple bottlenecks: heterologous enzyme incompatibility, metabolic flux imbalance, product toxicity, and economic viability. This review synthesizes recent breakthroughs in addressing these challenges through enzyme engineering, metabolic rewiring, host tolerance enhancement, and feedstock utilization. Simultaneously, the field is positioned at a critical juncture where convergent technologies — generative artificial intelligence for protein discovery, synthetic organelles via liquid–liquid phase separation LLPS, and engineering of non-natural terpenoid scaffolds (C₁₁, C₁₆) — promise transformative advances. This review provides a roadmap integrating these emerging capabilities to advance terpenoid-based biofuels toward commercial viability.

|
Scooped by
mhryu@live.com
January 23, 5:15 PM
|
Xanthan gum, a natural heteropolysaccharide produced by Xanthomonas species, has many biotechnological applications across industries due to its unique rheological properties. Expanding its utility requires specific enzymes capable of targeted xanthan modification or degradation. In this study, a novel bacterial strain, isolated from a spoiled xanthan sample and identified as Paenibacillus taichungensis I5, was shown to degrade xanthan using a plate screening assay with Congo red. Activity tests of crude enzyme in culture supernatant demonstrated the secretion of xanthan-degrading enzymes. Genome and proteome analyses suggest a chromosomal xanthan utilization locus encoding a suite of enzymes, including a xanthanase (Pt_XanGH9), two xanthan lyases (Pt_XanPL8a and Pt_XanPL8b), two unsaturated glucuronidases, two α-mannosidases, as well as transport and regulator proteins. Functional characterization through recombinant protein expression and enzyme assays confirmed the functions of Pt_XanGH9, Pt_XanPL8a and Pt_XanPL8b on native xanthan and xanthan-derived oligosaccharides. The polysaccharide degradation products released by these enzymes were identified via LC–MS analysis and suggested two xanthan lyases with divergent cleavage preferences. In contrast to Pt_XanPL8a, Pt_XanPL8b is synthesized with an N-terminal signal peptide, yet both lyases were detected in cell-free supernatant during growth on xanthan. Based on the composition of the xanthan utilization gene cluster and preliminary enzyme characteristics, a working model for xanthan utilization by P. taichungensis I5 is proposed. Reaching a better understanding of bacterial xanthan degrading pathways and the enzymes involved may help to develop modified xanthan derivatives and xanthan degrading enzymes that align with the specific demands of various industrial process.

|
Scooped by
mhryu@live.com
January 23, 5:09 PM
|
In natural and engineered ecosystems, diverse species interact in complex ways to form highly efficient microecologies. One key orchestrator of these interactions is autoinducer-2 (AI-2), a signaling molecule that plays a crucial role in microbial community assembly, metabolic flux, and resilience to environmental disturbances. This review provides the systematic synthesis of AI-2’s dual structural dynamics (S-THMF-borate/R-THMF interconversion) and its context-dependent roles in mediating bacterial crosstalk. It also reveals the receptor diversity (such as LuxP and LsrB) of AI-2 in bacterial kingdom and the signal transduction mechanism. Systematically elaborated on AI-2’s regulation of cellular metabolic flux and its ability to autonomously exhibit a series of coordinated behaviors in response to environmental changes. The review explores the ramifications of AI-2 on bacterial community interactions in synthetic biology and natural ecosystems. The wide application of AI-2-mediated interspecific communication in various fields including host health, agriculture, industry and environmental ecology has also been widely discussed. Factors influencing AI-2 production are thoroughly examined, including internal factors such as strain specificity, cell density, growth form and the phenotypic heterogeneity. Additionally, external biological factors (such as nutritional status and environmental stress) and abiotic factors (aggregation, diffusion, and flow) are discussed in detail. By examining knowledge gaps in AI-2-mediated spatial heterogeneity and multi-QS system coordination, this work charts a roadmap for harnessing microbial communication in chemical engineering and environmental sustainability.

|
Scooped by
mhryu@live.com
January 23, 3:36 PM
|
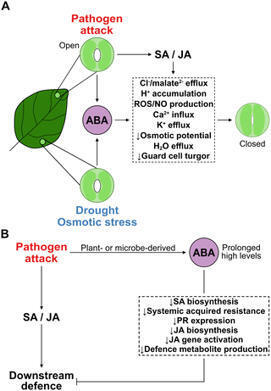
Abscisic acid sits at the nexus of abiotic and biotic stress signalling. It mediates fundamental responses to dehydration, salinity, and oxidative stress, while also influencing the outcome of pathogen encounters and symbiotic relationships with microbes. Through hormonal crosstalk, transcriptional regulation, and redox signalling, ABA fine-tunes stress responses and resource allocation. ABA signalling cross-talks with other hormonal pathways to balance growth, defence, symbiosis, and survival. Its effects are context- and ABA level-dependent, shaped by pathogen lifestyle, stress combinations, and tissue-specific dynamics. Decoding the full scope of ABA’s role in this crosstalk remains a key challenge. As climate change increases the frequency and complexity of stress combinations, understanding how ABA integrates a diversity of inputs will be essential for developing resilient crops and sustainable agricultural systems.

|
Scooped by
mhryu@live.com
January 23, 3:24 PM
|
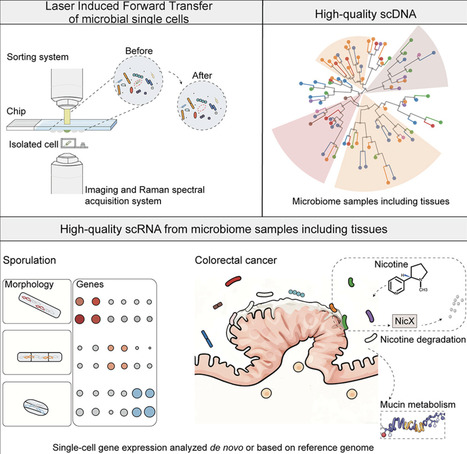
Metagenomics has enabled the understanding of the microbial composition and functional potential in various environments. Using laser-induced forward transfer (LIFT) technology, we report high-quality microbial single-cell genomes or transcriptomes in complex samples such as mouse gut, human saliva, and tumor sections. Bacterial cells in close proximity to each other or to host cells could be directly analyzed using this single-cell approach. Bacterial cells in mice or human samples could be fluorescently labeled for single-cell visualization before collection. The high-quality single-cell transcriptome results allow us to delineate cell-fate commitment in Bacillus sporulation and preliminary characterize gene expression from Bacteroides in a colorectal cancer sample. The method is scalable and precise and empowers insights about microbial populations and single-cell interactions with the host.

|
Scooped by
mhryu@live.com
January 23, 2:24 PM
|
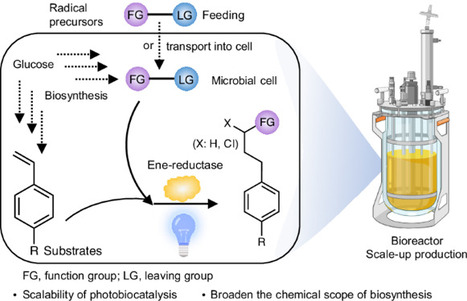
Photobiocatalysis provides a powerful strategy for integrating light and biological catalysts to drive abiological transformations. However, its scalability is hindered by high enzyme loading, reliance on costly cofactors and instability under radical-generating conditions. Here we report the integration of light-driven enzymatic reactions into the cellular metabolism of Escherichia coli, bridging flavin-based photobiocatalysis with biosynthesis. Using synthetic biology strategies, we engineered microbial cells to continuously produce olefin substrates and ene-reductase while regenerating cofactors directly from glucose. By externally supplying radical precursors or introducing synthetic pathways for their in situ production, we enabled fermentation-based microbial photobiosynthesis, achieving high titres and demonstrating feasibility for scale-up in a bioreactor. This approach extends photobiocatalysis from in vitro applications to in vivo semi- and complete biosynthesis, revealing its full potential for integrating light-driven reactions into cellular metabolism. Light-driven enzymatic catalysis has enabled important abiological transformations in vitro. Now a cellular ene-reductase photoenzyme is integrated with a de novo-designed olefin biosynthetic pathway for photoinduced hydroalkylation, hydroamination and hydrosulfonylation reactions within cells.

|
Scooped by
mhryu@live.com
January 23, 1:55 PM
|
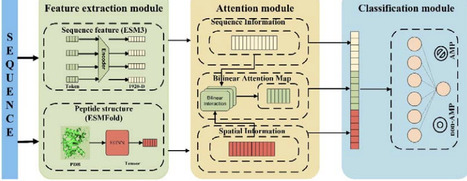
The escalating crisis of antimicrobial resistance poses a devastating and immediate threat to human life. Antimicrobial peptides (AMP) are a promising antibiotic substitute to combat antimicrobial resistance. Compared with the traditional wet-lab screening approaches, computational models have largely improved the efficiency of predicting antimicrobial peptide. However, most computational models overlook or underutilize the evolution and structural information of peptides, which is crucial for understanding the peptide functions. Here, we proposed a sophisticated deep learning model to predict AMPs, Antimicrobial Peptide Bilinear Attention Network (AMPBAN), which incorporates peptide evolution features from ESM3 protein language model, structure features from ESMFold predicted with equivariant graph neural network (EGNN), and the joint information from sequence and structure learned via Bilinear Attention Network. AMPBAN consistently demonstrated superior accuracy and generalization compared to nine state-of-the-art AMP prediction models across multiple independent benchmarks. Furthermore, an ablation study confirms that our multimodal fusion strategy significantly refines the integration of sequence and structural signals, yielding superior predictive balance over single-modality models. This framework provides a robust tool for the accelerated discovery of novel AMPs and the advancement of next-generation antimicrobial drug development. The datasets, source code and models are available at https://github.com/baiwenhuim/ampban.

|
Scooped by
mhryu@live.com
January 23, 1:31 PM
|
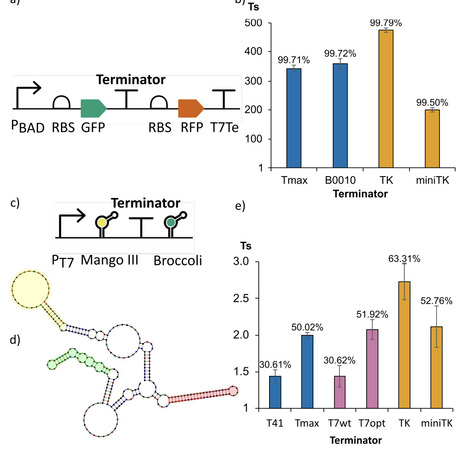
Intrinsic transcription terminators are biological parts critical for controlling gene expression in natural genomes and are fundamental to the modularity and predictability of synthetic gene circuits. Despite their simplicity of structure and function, we have not yet been able to rationally engineer synthetic terminators with a pre-defined strength, nor to accurately predict their strength from sequence. Here, we leveraged a curated library of bacterial terminators to train a data-driven predictive model, and, building on this surrogate, developed open-source software tools for predicting terminator performance and designing new intrinsic terminator sequences. Model interpretability analysis indicates that U-tract features emphasize a distal region longer than previously anticipated and that the initial hairpin GC content influence extends beyond the reported range. Using the final trained model, we implemented two software tools. The Terminator Strength Predictor (TerSP) computes the full feature representation directly from an input sequence and outputs a quantitative strength prediction together with a binary strong/weak classification. We validated TerSP using experimentally characterized terminators from bacteria other than E. coli. The Terminator Factory (TerFac) implements a surrogate-based optimization framework for target-driven terminator design under user-defined strength and length constraints. Using TerFac, we enumerated length-specific sets of maximally strong terminators, designed optimized synthetic terminators, and optimized a wild-type terminator. The designed terminators were validated in vivo in E. coli and in vitro, using a newly developed assay based on fluorescent RNA aptamers. The TerFac-designed terminators showed the expected strength, and the strongest one outperformed the best reference terminator in the training dataset, both in vivo and in vitro. These results indicate that the model captured sequence-to-function rules that are informative both for forward prediction (TerSP) and for the design of terminators with defined strength (TerFac).

|
Scooped by
mhryu@live.com
January 23, 1:16 PM
|
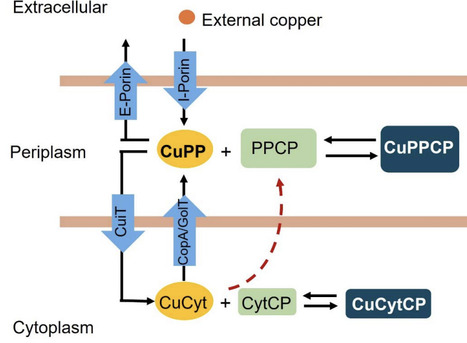
Copper (Cu) is an essential micronutrient that serves as a cofactor for redox enzymes but becomes toxic when unregulated. In bacteria, while Cu efflux systems are well characterized, mechanisms of Cu import remain poorly understood. Here, we characterize the major facilitator superfamily transporter CuiT (STM1486) as a key Cu importer in Salmonella enterica. Comparative genomics revealed that cuiT is evolutionarily conserved across Enterobacteriaceae, and structural modeling predicts a 12-transmembrane-helix architecture with conserved His, Met, and Cys residues suitable for Cu⁺ coordination. Functional analyses demonstrated that deletion of cuiT reduces intracellular Cu accumulation, slows Cu uptake kinetics, and diminishes expression of Cu-responsive genes, including copA, cueP, cueO, and golB. Conversely, overexpression of CuiT increases intracellular Cu but sensitizes cells to Cu stress, highlighting the need for tight regulation. Kinetic modeling indicates that CuiT mediates rapid Cu import, supporting larger intracellular Cu pools compared to Pseudomonas influx transporters. These findings position CuiT as a central component of the Salmonella Cu homeostasis network, linking Cu import to transcriptional regulation, redox balance, and stress adaptation. Our work provides mechanistic insights into bacterial Cu acquisition and suggests CuiT and associated pathways as potential targets for antimicrobial strategies.

|
Scooped by
mhryu@live.com
January 23, 10:43 AM
|
The gut microbiome plays an active role in host health, producing gut metabolites that influence host digestive and immune function while also mediating microbial crosstalk. Dietary fiber is a major source of important fermentation byproducts that are generally implicated in gut community stability and host wellbeing, but dissecting microbe-specific contributions to polysaccharide metabolism in the context of a complex gut community is challenging with conventional model organisms. Using the American cockroach (Periplaneta americana) as a model omnivore, we use chemically-defined synthetic diets to identify how complex gut microbial communities respond to two of the most abundant plant polysaccharides, xylan and cellulose. To do so, we fed cockroaches synthetic diets containing one of these fibers or a mix of both in differing ratios. Through both 16S rRNA gene profiling and RNA-seq, we show that mixed fibers enrich for organisms characteristic of the source fibers as well as additional organisms only enriched in mixed-fiber diets. Through an organism-centric pangenome approach, we identify the impact of these fibers on gut microbiome activity. We found that gut communities responded strongly to xylan, with Bacteroidota belonging to Bacteroides, Dysgonomonas, and Parabacteroides producing xylan-active CAZymes at high levels. Multiple groups of Bacillota also responded strongly to a xylan diet, but appeared to act as cross-feeding secondary degraders, producing primarily xylosidases and transcripts associated with xylose utilization. In contrast, cellulose diets were associated with higher transcriptional activity among Fibrobacterota, which are typically a minor component of the cockroach gut microbiome but were the primary producers of CAZymes associated with cellulose and cellobiose degradation. These experiments provide new insight into gut microbial metabolism of these complex plant polysaccharides. Further, they highlight the utility of the cockroach model and synthetic diets to answer fundamental questions about gut microbial responses to different polysaccharides alone and in combination.

|
Scooped by
mhryu@live.com
January 23, 9:47 AM
|
The root microbiome plays a critical role in nutrient acquisition, stress tolerance and overall plant health. Rice, a staple food for more than half of the population of the world, is commonly cultivated under flooded conditions. Despite its agronomical importance, our current understanding of root-associated microbiomes in rice grown under flooded conditions is limited. On the other hand, nitrogen and phosphorus fertilizers are routinely applied to maximize rice yield. It is also well known that root colonization by arbuscular mycorrhizal fungi enhances mineral nutrition in plants, but whether mycorrhizal associations influence the composition of the rice root microbiome remains poorly understood. In this study, shotgun metagenomic sequencing was used to characterize the root endosphere and rhizosphere microbiomes in two temperate japonica rice varieties (cv. Bomba and JSendra) grown under flooded conditions. The impact of colonization by the AMF Rhizophagus irregularis on the root microbiome was investigated. Root-associated compartments harbor distinct microbial communities in rice with bacterial taxa comprising approximately 95% of the total microbia in rice roots. At the Phylum level, the root bacteriome was primarily composed of Pseudomonadota (Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria) followed by Actinomycetota. The fungal microbiome was dominated by Ascomycota (Sordariomycetes, Eurotiomycetes and Dothideomycetes) and Basidiomycota. Not only the root compartment, but also the host genotype can shape the root microbiome. Recruitment of specific microorganism mainly occurs at the species level. Genotype-specific and compartment-specific associations of microbial species in mycorrhizal rice roots were also observed supporting that root colonization by an AMF contributes to variations in the root microbiome. Further, key microbial species primarily associated to methane production and nutrient cycling (e.g., Phosphate Solubilizing Bacteria and Nitrogen cycling bacteria) colonizing root compartments in each rice genotype and mycorrhizal condition are described.

|
Scooped by
mhryu@live.com
January 23, 9:30 AM
|
Archaeal extrachromosomal elements (ECEs) are arguably the least well understood of all genetic elements, and few have > 200 kbp ("jumbo") genomes. Here, we report circular, jumbo ECEs with genomes of up to 535 kbp in length that associate with anaerobic methane-oxidizing Methanoperedens archaea. Notably, a 409-kbp genome related to jumbo ECEs is integrated into a subset of the ~4.2 Mbp Methanoperedens chromosomes at the tRNA-Asp genes. This represents the largest integrative element in Archaea and supports the jumbo ECE - host association. Multiple genome alignment and phylogenetic analyses suggest that the large sizes were developed by extensive DNA acquisition from Methanoperedens. The newly identified ECEs encode, and in some cases express, metabolic genes such as tetrahydromethanopterin S-methyltransferase exclusively involved in methane oxidation, and genes for nitrogen and sulfur compound transformations. Also encoded are defense systems, some of which are absent in hosts, such as hybrid Type I/Type III-A CRISPR-Cas systems. In contrast to viruses and plasmids, they have host-like replication machinery and occur at stable copy ratios of 1.44 ± 0.24 : 1 to the host. Overall, our results reveal a spectrum of jumbo ECEs of Methanoperedens, ranging from plasmid-like to minichromosome-like.
|

|
Scooped by
mhryu@live.com
January 23, 5:26 PM
|
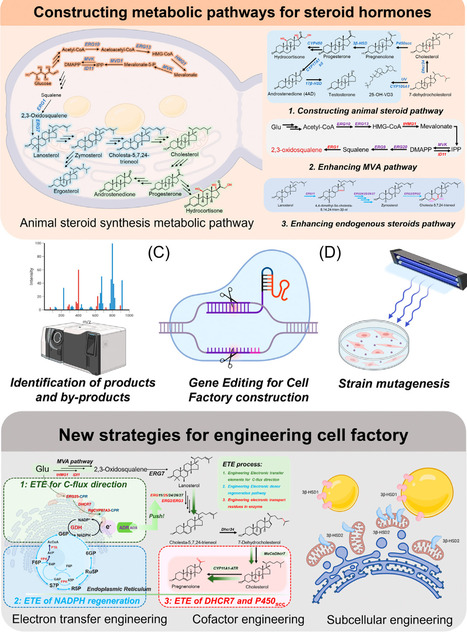
Steroid hormones are key signaling molecules regulating growth, metabolism, reproduction, and stress adaptation and are widely used as essential pharmaceuticals. Traditional production from sterol feedstocks through multistep chemical or microbial transformations is limited by inefficiency and scalability. Recent advances in synthetic biotechnology enable de novo biosynthesis of steroid hormones from simple carbon sources in yeasts and fungi. This review highlights metabolic rewiring to increase flux, cytochrome P450 enzyme engineering for side-chain cleavage, and hydroxylation to overcome rate-limiting bottlenecks of steroid hormone biosynthesis. We also discuss strategies to redesign steroid-transport pathways to alleviate intracellular accumulation and improve membrane export. Looking ahead, we envision integrating metabolic, enzyme, and transport engineering to build a scalable, data-driven ‘intelligent’ platform for sustainable steroid hormone biomanufacturing.

|
Scooped by
mhryu@live.com
January 23, 5:17 PM
|
Development of chemically modified oligonucleotides, nucleic acid mimics, protein-based constructs, and other ligands -capable of sequence-unrestricted recognition of specific double-stranded (ds) DNA regions -is an area of research that continues to attract considerable attention. Efforts are fueled by the need for diagnostic agents, modulators of gene expression, and novel therapeutic modalities against genetic diseases. While pioneering approaches focused on accessing nucleotide-specific features from the grooves of DNA duplexes, recent developments have entailed strand-invading probes, i.e., probes capable of binding to DNA duplexes by breaking existing Watson-Crick base pairs and forming new, more stable base pairs. For the past twenty years, our laboratory has pursued the development of a type of dsDNA-targeting strand-invading probes, which we have named Invader probes. These double-stranded oligonucleotide probes feature intercalator-functionalized nucleotides that are specifically arranged to promote destabilization of the probe duplex, whereas individual strands exhibit very high affinity towards complementary DNA. This account details the discovery, principles, and applications of Invader probes.

|
Scooped by
mhryu@live.com
January 23, 5:13 PM
|
Virus-induced genome editing (VIGE) using compact RNA-guided endonucleases is a transformational new approach in plant biotechnology, enabling tissue-culture-independent and transgene-free genome editing. We recently established a transgene-free VIGE approach for heritable editing at single loci in Arabidopsis by delivering ISYmu1 TnpB (Ymu1) and its guide RNA (gRNA) via Tobacco Rattle Virus (TRV). Here, we greatly improved this approach by devising a multiple gRNA expression system and by utilizing an engineered high-activity Ymu1 variant (Ymu1-WFR) to develop an efficient multiplexed genome editing approach.

|
Scooped by
mhryu@live.com
January 23, 3:43 PM
|
Effectors are diverse molecules produced and secreted by plant pathogens to facilitate infection often by exerting an “effect” on their host. The production of effector molecules is a ubiquitous feature of plant pathogens. Because many effector proteins act on host targets and are subject of host immune detection, the corresponding effector genes co-evolve with the corresponding host genes involved in immunity or susceptibility. This co-evolution results in diversity at effector loci, both within and between lineages/species. Here, we examine the diverse patterns and consequences of variation observed across the sequence and regulatory landscapes of effector genes and summarize the genomic mechanisms that create them.

|
Scooped by
mhryu@live.com
January 23, 3:31 PM
|
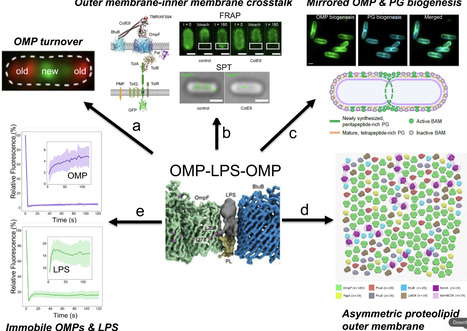
Bacteriocins are toxins deployed by bacteria to kill their competitors. Here, I reflect on my laboratory’s work on protein bacteriocins and their immunity proteins from Gram-negative bacteria. We uncovered the structural and biophysical principles that underpin the ten-log stability range of protective immunity proteins for cytotoxic nuclease bacteriocins. We went on to elucidate how bacteriocins that kill Escherichia coli, Pseudomonas aeruginosa and Klebsiella pneumoniae subvert outer membrane proteins and periplasmic energy transduction systems to drive their import. We leveraged our understanding of bacteriocin structure and function to probe the nature of the bacterial outer membrane. These studies revealed the outer membrane of E. coli to be an asymmetric proteolipid membrane, not an asymmetric lipid membrane as has been accepted dogma for the last 50 years. I contextualise the work with background on my career, collaborations and academic leadership roles that influenced my development as a scientist.

|
Scooped by
mhryu@live.com
January 23, 3:19 PM
|
The emergence of antibiotic-resistant bacteria has rendered conventional antibiotic treatments ineffective, necessitating the development of antibacterial agents with unique mechanisms of action. To address this challenge, researchers have increasingly resorted to synthetic and bioengineered nano-materials to augment the antibacterial activity of nonantibiotic antibacterials (nonantibiotic antibacterial agents), including antimicrobial peptides (AMPs), metallic nanoparticles (MNPs), bacteriophages (phages), and phage derivatives such as endolysins, which are under extensive investigation. In this review, we discuss how modifications and syntheses of these agents, leveraging advancements in nanoscience and nanotechnology, have and can significantly enhance their antibacterial properties and overcome limitations such as cytotoxicity, instability, and poor bioavailability for in vivo or clinical use. Furthermore, we highlight supramolecular strategies for improved delivery, including phage-based, AMP-based, and endolysin-based systems and their demonstrated efficacy against persistent bacterial infections. Additionally, we highlight how the integration of artificial intelligence and machine learning ultimately promises to revolutionize the design, optimization, and clinical translation of these precision antimicrobials, paving the way for targeted and highly effective treatments.

|
Scooped by
mhryu@live.com
January 23, 2:19 PM
|
Plant diseases caused by Pseudomonas syringae pathovars pose a substantial threat to global crop production. While plants produce a range of secondary metabolites as part of their defense response, how bacterial pathogens exploit these compounds to facilitate infection remains poorly understood. Here, we report that Pseudomonas syringae pv. actinidiae (Psa), the causal agent of kiwifruit bacterial canker, senses putrescine (Put), a crucial plant metabolite involved in defense and physiological regulation, via the histidine kinase BvgS. We identify BvgS as a Put receptor in bacteria and show that this perception upregulates the expression of the type III secretion system (T3SS), a major virulence determinant. Binding and virulence assays demonstrate that Put sensing through the PBPb domains of BvgS is essential for T3SS activation both in vitro and during plant infection. Modulating Put levels in Actinidia plants, either by genetic reduction or exogenous application, correspondingly alters T3SS activity and bacterial invasion. Evolutionary analysis indicates that the PBPb domain is highly conserved across diverse P. syringae pathovars, suggesting a widespread mechanism for virulence potentiation. Together, this work delineates a signaling pathway through which a phytopathogen co-opts a central host metabolite to enhance the expression of its core virulence machinery, thereby increasing infectivity.

|
Scooped by
mhryu@live.com
January 23, 1:50 PM
|
Metagenome-assembled genomes (MAGs) are routinely recovered from metagenomic studies, yet the population genetic information embedded within these datasets remains largely underutilized. Analyzing within-species genetic variation can reveal adaptive evolution, selection pressures, and ecological dynamics that are hidden when MAGs are treated as homogeneous entities. Existing tools address individual analysis steps in isolation, requiring manual integration and creating barriers for researchers without extensive bioinformatics expertise. Here we present PopMAG, a Nextflow pipeline and interactive Shiny application that automates population genetics analysis of MAGs. PopMAG integrates quality control, community profiling, competitive read mapping, functional annotation, and microdiversity estimation into a single reproducible workflow. The pipeline calculates key population genetics metrics including nucleotide diversity (π), pN/pS ratios, fixation index (FST), Levins index and SNVs counts with results consolidated into an interactive visualization platform for metadata-driven exploration. We demonstrate PopMAGs utility through analysis of longitudinal cystic fibrosis lung metagenomes, where we detect signatures of antibiotic-driven selection in Pseudomonas aeruginosa efflux pump genes coinciding with treatment intervention. Availability and implementation: PopMAG and corresponding documentation are publicly available at https://github.com/daasabogalro/PopMAG

|
Scooped by
mhryu@live.com
January 23, 1:22 PM
|
A methodology is presented to integrate DNA methyltransferase genes of interest into the single copy, IncPbeta; R Factor conjugal plasmid, R702. This is to enable DNA transfer, or improved frequency thereof, by mirroring the recipient native DNA methylation signature prior to transfer of plasmid DNA by conjugation, evading native restriction modification systems in the organism of study, without target strain modification. As proof of concept, the bacteriophage derived methyltransferase phi3TI, known to facilitate DNA transfer into the industrially important model solventogenic organism, Clostridium acetobutylicum ATCC 824, was inserted into the single copy R702 to create a methylating strain. Plasmids extracted from this strain were transferred by electroporation at a comparable frequency to the established method using the multi-copy accessory plasmids pAN1 or pAN2, harbouring phi3TI. The methodology was further exemplified and employed to enable high frequency of DNA transfer into the clinically relevant Clostridioides difficile ribotype 027 outbreak strain, R20291. The modification (hsdM) and specificity (hsdS) components of the native Type I restriction modification system of R20291 was inserted into R702 to create a functional methylating conjugal donor Escherichia coli strain. This approach of coupling methylation and the native transfer functions of R702 creates a system which can easily be mobilised into a genotypically appropriate E. coli strain; creating an in-vivo methylating donor strain which may be utilized to protect and transfer DNA to the organism of study by conjugation.

|
Scooped by
mhryu@live.com
January 23, 1:14 PM
|
Oxygen is a primary driver of the distribution and activity of microbial life. Since oxygen levels are often difficult to measure in situ, one potential solution is to use bacteria as bioindicators of oxygen levels. As bacteria range from obligate aerobes to obligate anaerobes, quantification of bacterial community oxygen preferences could be used to infer variation in environmental oxygen levels and bacterial metabolic strategies. After using ensemble machine learning to select the 20 most important genes that predict oxygen tolerances in individual bacteria, we established a relationship between the abundance ratio of aerobic: anaerobic indicator genes and the proportional abundance of aerobic bacteria using simulated metagenomes with varying ratios of known aerobic and anaerobic bacteria. We developed a tool, OxyMetaG, that takes metagenomic reads as input, extracts bacterial reads, maps reads to the 20 genes, and predicts the proportion of aerobic versus anaerobic bacteria in any given sample. We tested OxyMetaG on a suite of metagenomes with measured or inferred oxygen levels across a variety of environmental and host-associated samples. To demonstrate the utility of our approach, we applied OxyMetaG to 540 surface soils, showing that surface soils are typically dominated by aerobes, but wetter sites with finer textures have relatively more anaerobes. Lastly, we applied OxyMetaG to 73 human gut samples, showing that in the first three years of life, human guts progress from having up to 61% aerobes to being completely dominated by anaerobes. We expect OxyMetaG to have broad utility for characterizing both modern and ancient environments.

|
Scooped by
mhryu@live.com
January 23, 9:56 AM
|
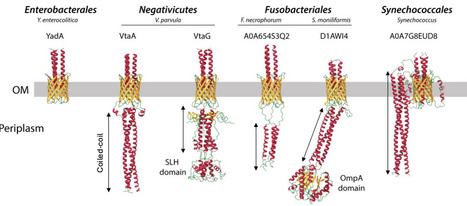
Autotransporters are important gram-negative bacterial cell-surface proteins and are virulence factors enabling surface attachment and adhesion to other bacteria. These proteins are composed of a signal peptide, a β-barrel that serves as an anchor in the outer membrane, and an extracellular passenger domain responsible for adhesion. Autotransporters rely on BamA for their insertion in the outer membrane (OM), but specific helper proteins, such as TpgA and SadB have been described to promote the surface exposure of trimeric autotransporters (TAAs), a specialized subclass of autotransporters forming homotrimer adhesins. To identify domains or proteins that could help TAAs secretion, we analyzed a recent dataset of all trimeric autotransporters found across the bacterial tree of life. While we did not find additional potential helper proteins for OM translocation, we found that the extended signal peptide (ESPR), sometimes found in TAAs, is associated with longer adhesins both for TAAs and type Va autotransporter adhesins. ESPRs are found in all bacteria but Fusobacteriia and Alphaproteobacteria. We also identified in Burkholderia, Veillonellales and Pasteurellales a DUF2827 domain proteins as potential glycosyltransferases constantly associated with TAAs. Finally, we describe the existence of extra periplasmic domains in some TAAs, featuring either a coiled-coil domain or a peptidoglycan-binding domain. Our research show that there is a strong phylogenetic separation between Terrabacteria, almost invariably displaying additional periplasmic domains, and Gracilicutes (represented by Proteobacteria) where they are largely absent. This suggests that the presence these domains might be correlated with specific Terrabacteria OM features. Using the diderm Firmicute Veillonella parvula as a model, we demonstrate that the absence of periplasmic domains in TAAs leads to a significant protein degradation, yet they are not essential for adhesin trimerization or secretion. Additionaly, we show that the SLH domains of V. parvula TAAs excludes them from the septum during division, but that this exclusion is not crucial for adhesin function or stability in the tested conditions. Altogether, these results illuminate the genetic flexibility and modularity of autotransporters, enhancing our understanding of this important class of adhesins in diderm bacteria.

|
Scooped by
mhryu@live.com
January 23, 9:43 AM
|
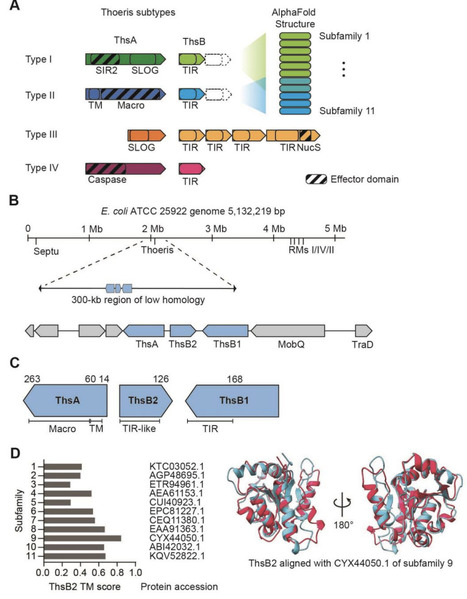
Thoeris immune systems protect bacteria against invading phages through production of a signaling molecule by ThsB that activates the effector ThsA. Across the four defined types, the two most dominant (I and II) have been associated with abortive infection, with type I acting through the depletion of the essential coenzyme NAD+. Here, we show that a previously uncharacterized type II Thoeris system from Escherichia coli ATCC 25922 deviates from this paradigm. This system consists of the effector protein ThsA and two distinct signaling proteins ThsB1 and ThsB2. Heterologous expression of thsA and thsB2 confers antiphage defense, while thsB1 is cytotoxic when expressed in common E. coli lab strains. Furthermore, while phage infection drives growth arrest, we could not detect any measurable decrease in NAD+ levels as well as standard markers of cell death. Together, these results suggest that Thoeris contains even greater functional diversity within the defined system types.
|
 Your new post is loading...
Your new post is loading...