Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 7:26 PM
|
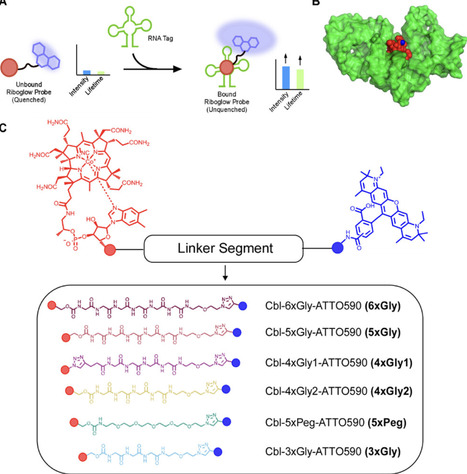
Riboglow probes are small molecules where a synthetic fluorophore is connected to an RNA-binding moiety via a chemical linker. Upon binding a short RNA sequence, probe fluorescence intensity and lifetime increase. The fluorescence change is modulated by the architecture of the chemical linker. Here, we systematically interrogated the linker composition in a series of Riboglow probes and assessed fluorescence properties. We found that glycine linkers result in higher fluorescence turn-on compared to a polyethylene glycol linker of similar length. When varying the length of the polyglycine linker, we found that increasing the number of glycine residues led to more substantial fluorescence turn-on upon RNA-ligand binding. Surprisingly, the composition of the Riboglow chemical linker influences fluorescence lifetime contrast when comparing probe binding to two different RNA ligands, a quality necessary for RNA multiplexing. Finally, evaluating probe fluorescence lifetimes in live mammalian cells demonstrated the ability of new Riboglow probes to visualize RNAs live. Insights gained from the systematic assessment of the linker’s architecture will dictate the rational design of future fluorophore-quencher probe designs.

|
Scooped by
mhryu@live.com
Today, 4:58 PM
|
Antibiotic resistance (AR) is an escalating public health threat, necessitating innovative strategies to control resistant bacterial populations. One promising approach involves engineering genetic elements that can spread within microbial communities to eliminate AR genes. Previously, we developed Pro-Active Genetics (Pro-AG), a CRISPR-based gene-drive-like system capable of reducing AR colony-forming units (CFU) by approximately five logs. Here, we advance this technology by integrating Pro-AG into a conjugative transfer system, enabling efficient dissemination of an anti-AR gene cassette between two bacterial strains. Additionally, we characterize a complementary homology-based deletion (HBD) process, a CRISPR-driven mechanism that precisely removes target DNA sequences flanked by short direct repeats. Our findings reveal that Pro-AG and HBD are differentially influenced by the bacterial RecA pathway and that HBD components can be delivered via plasmids or phages to selectively delete Pro-AG cassettes. This built-in safeguard prevents uncontrolled spread of a gene cassette and mitigates unanticipated side effects. These refinements enhance the efficiency and flexibility of Pro-AG, expanding its potential applications in microbiome engineering, environmental remediation, and clinical interventions aimed at combating antibiotic resistance. More broadly, this work establishes a proof-of-principle for microbiome engineering strategies that could be leveraged to improve health and restore ecological balance.

|
Scooped by
mhryu@live.com
Today, 4:46 PM
|
The increasing imperative to mitigate greenhouse gas emissions and foster the transition to a low-carbon bioeconomy has intensified interest in methane bioconversion as a sustainable approach for transforming methane into valuable bioproduction. Although advancements have been made in optimizing methanotrophic pathways to improve bioproduction, significant challenges persist, including methane solubility, bioavailability, and metabolic flexibility, limiting the efficiency of methane conversion. This review provides a comprehensive overview of the initiatives aimed at developing next-generation methanotrophic cell factories by overcoming the physiological limitations of natural methanotrophs. We first analyze the metabolic characteristics of methanotrophs for assimilating methane into cellular building blocks. Then, we discuss methane assimilation pathways and their unique characteristics in matter and energy transmission for facilitating the integration of methane into central carbon metabolism. Further, we propose a systematic framework for designing methane-based biomanufacturing to enable low-carbon bioproduction by integrating synthetic biology, metabolic engineering, and systems biology, thereby developing efficient methane assimilation cell factories for producing high-value bioproducts. Finally, we prospect the potential for valorizing methane derived from anthropogenic emissions and renewable sources, while identifying the key challenges and future research directions necessary for advancing a sustainable, low-carbon bioeconomy.

|
Scooped by
mhryu@live.com
Today, 4:31 PM
|
Antibiotic resistance is escalating, highlighting the urgent need for novel antimicrobial strategies. Defensin-like antimicrobial peptides (AMPs) are considered ideal candidates due to their broad-spectrum activity and engineerable potential; however, their limited antimicrobial efficacy and complex chemical synthesis constrain practical applications. In this study, we aimed to enhance the antimicrobial properties of defensin-like AMP through rational design, directed evolution, and structural fusion strategies. The engineered variant XC1 demonstrated significantly improved antimicrobial activity against a broad range of pathogens, including methicillin-resistant Staphylococcus aureus, while maintaining broad-spectrum efficacy. Comprehensive evaluation of toxicity and stability showed that XC1 exhibited good functional stability in serum, low hemolysis, and low cytotoxicity, indicating excellent therapeutic potential. In addition, high-level secretory expression of defensin-derived AMPs and their engineered variants was achieved using Pichia pastoris GS115, demonstrating strong biosynthetic capability. Together, these results provide a viable strategy for enhancing the antimicrobial activity and scalable biosynthesis of defensin-like AMPs.

|
Scooped by
mhryu@live.com
Today, 4:14 PM
|
Carotenoids and apocarotenoids constitute a structurally and functionally sundry class of isoprenoids whose significance extends from photosynthetic light capture and photoprotection to phytohormone signaling, flavor and aroma formation, and emerging biomedical applications. While recent appraisals have emphasized quantitative advances in microbial production, this mini-review adopts a pathway module-centric perspective. We examine each biosynthetic stage from precursor supply, condensation to geranylgeranyl diphosphate (GGPP), phytoene synthesis, desaturation/isomerization, cyclization, hydroxylation, ketolation, epoxidation, and oxidative cleavage, highlighting novel enzymatic variants, mutagenesis studies, fusion strategies, and compartmentalization approaches that impart metabolic control. Special emphasis is placed on recently discovered and engineered enzymes, as well as synthetic biology tools. This review integrates diverse enzyme sources, host ranges across plants, fungi, algae, yeasts, and bacteria, as well as pathway modularity, to provide an updated review of recent literature. We conclude by outlining future directions that highlight gaps and potential areas for future work. This focused synthesis aims to equip researchers with a hierarchical understanding of the pathways and strategies to advance carotenoid and apocarotenoid biosynthesis.

|
Scooped by
mhryu@live.com
Today, 2:25 PM
|
The engineering of enzymes with novel functions is a cornerstone of synthetic biology but remains bottlenecked by the fragmentation between computational design and physical execution. While "self-driving" laboratories promise to resolve this, existing systems often rely on rigid, device-specific scripts that lack the flexibility to handle complex, evolving scientific tasks. Here, we report an AI-native autonomous biofoundry that fundamentally redefines laboratory automation through a "cloud-edge synergistic" architecture. The platform features an Agent-Native control system powered by Large Language Models (LLMs) and the Model Context Protocol (MCP), which bridges the semantic gap between abstract scientific intent and heterogeneous hardware execution. This architecture enables non-experts to orchestrate the entire Design-Build-Test-Learn (DBTL) cycle via natural language. By integrating deep phylogenetic mining, zero-shot protein language models (ESM-2), and supervised active learning, our system efficiently navigates rugged fitness landscapes. As a rigorous proof of concept, we applied this platform to evolve a Family B DNA polymerase for CoolMPS sequencing, a task requiring the incorporation of non-natural 3'-blocked nucleotides. In just three autonomous rounds, the platform achieved a hit rate of >66% and identified variants with a 37% reduction in sequencing error rate compared to a commercial reference. This work demonstrates that AI-native infrastructures can not only accelerate trait evolution by orders of magnitude but also provide a scalable, brand-agnostic paradigm for the future of automated scientific discovery.

|
Scooped by
mhryu@live.com
Today, 2:10 PM
|
Mixotrophy offers a promising strategy for biosynthesis by simultaneously utilizing organic carbon and CO2; however, mixotrophic microorganisms are rarely isolated outside of photoautotrophic microalgae. In this study, the chemoautotroph Cupriavidus necator H16 was found to preferentially consume fructose when coexisting with CO2 and H2, switching to utilization of only CO2 and H2 after fructose depletion. Transcriptomic analysis revealed significant differences in genes involved in energy metabolism, electron generation, and the respiratory chain. The molecular mechanism underlying the inability of C. necator H16 to simultaneously utilize carbohydrates and CO2 was identified as the suppression of hydrogenase expression in the presence of fructose. By inducing regulator hoxA to activate hydrogenase expression, an engineered C. necator strain capable of mixotrophic growth was developed. This engineered strain can simultaneously utilize fructose, CO2, and H2, maintain optimal growth, and approach carbon-neutral cultivation. This work provides insights for the mixotrophic cultivation of C. necator and serves as a reference for developing mixotrophic microorganisms in future studies.

|
Scooped by
mhryu@live.com
Today, 12:48 PM
|
In modern agriculture, microbial inoculants isolated and collected from all over the world have gained popularity as a means of reducing the amount of fertilizer by increasing the availability of nutrients and mitigating environmental stress that is often connected with climate change. Concerning biocontrol, microbial inoculants are known to be effective in integrated pest management. However, the introduction of alien microbes can lead to the emergence of antagonists of the natural soil microbiota, which might drastically change the latter and ultimately have a negative impact on the whole natural soil ecosystem, causing unforeseeable consequences. We will discuss various aspects of the employment of microbial inoculants in agriculture, with a focus on the largely neglected threat posed by potentially invasive microbes.

|
Scooped by
mhryu@live.com
Today, 12:26 PM
|
Nucleic acid sequencing is the process of identifying the sequence of DNA or RNA, with DNA used for genomes and RNA for transcriptomes. Deciphering this information has the potential to greatly advance our understanding of genomic features and cellular functions. In comparison to other available sequencing methods, nanopore sequencing stands out due to its unique advantages of processing long nucleic acid strands in real time, within a small portable device, enabling the rapid analysis of samples in diverse settings. Evolving over the past decade, nanopore sequencing remains in a state of ongoing development and refinement, resulting in persistent challenges in protocols and technology. This article employs an interdisciplinary approach, evaluating experimental and computational methods to address critical gaps in our understanding in order to maximize the information gain from this advancing technology. Here, we review both overview and analysis of all aspects of nanopore sequencing by providing statistically supported insights. Thus, we aim to provide fresh perspectives on nanopore sequencing and give comprehensive guidelines for the diverse challenges that frequently impede optimal experimental outcomes.

|
Scooped by
mhryu@live.com
Today, 11:38 AM
|
Suppressor transfer RNAs (sup-tRNAs) have the potential to rescue nonsense mutations in a disease-agnostic manner and are an alternative therapeutic approach for many rare and ultrarare disorders. Among all human pathogenic nonsense variants, approximately 20% arise from C-to-T transitions that convert the CGA arginine codon into a UGA stop codon. While recombinant adeno-associated virus (rAAV) has been successfully used to deliver a UAG-targeting sup-tRNA gene in vivo, extending this approach to UGA-targeting sup-tRNA genes has posed unique challenges related to rAAV vector production. Here, we demonstrate that an engineered UGA-sup-tRNA gene, designed with transcriptional regulatory elements, can be efficiently packaged into rAAV for in vivo delivery. A single administration in mouse models of two distinct lysosomal storage disorders restores enzymatic activity to approximately 10% of normal levels. Comparative analysis reveals differential sup-tRNA expression and aminoacylation patterns across tissue types, which correlate with enhanced therapeutic effects. The applied rAAV-based agents and engineering strategies expand the potential therapeutic scope of sup-tRNA therapies. Therapeutic suppressor tRNAs are engineered to broaden their target range in vivo.

|
Scooped by
mhryu@live.com
February 1, 1:35 PM
|
MicroRNAs (miRs) are small non-coding RNAs that regulate gene expression. Their dysregulation is closely associated with various diseases, positioning them as biomarkers of cellular state. Synthetic biology has leveraged these properties to engineer miR-based genetic circuits capable of sensing and interpreting endogenous miR levels. Early miR-OFF systems relied on reporter gene repression but were limited by ambiguous signal interpretation. Subsequent advances introduced miR-ON architectures, logic-based classifiers integrating multiple miRs, and layered regulatory strategies combining transcriptional, translational, and cleavage-based modules to enhance sensitivity and specificity. Recent innovations include CRISPR-associated miR-responsive systems and incoherent feed-forward loop (iFFL) architectures that stabilize gene expression amid cellular variability, shifting applications from passive sensing to therapeutic intervention. Despite challenges such as leakage, cellular resource resources, and delivery, progress in orthogonal miR toolkits, computational modeling, and RNA-based delivery platforms is rapidly driving miR-based circuits toward diagnostic and therapeutic applications.

|
Scooped by
mhryu@live.com
February 1, 1:24 PM
|
Invasive fungal infections claim over two million lives annually, a problem exacerbated by rising resistance to current antifungal treatments and an increasing population of immunocompromised individuals. Despite this, antifungal drug development has stagnated, with few novel agents and fewer novel targets explored in recent decades. Here, we validate acetolactate synthase (ALS), an enzyme critical for branched-chain amino acid biosynthesis and absent in humans, as a promising target for new therapeutics. Using resistance gene-guided genome mining, we discovered a biosynthetic gene cluster BGC in Aspergillus terreus encoding HB-35018 (1), a novel spiro-cis-decalin tetramic acid that potently inhibits ALS. Biochemical and antifungal assays demonstrate that 1 surpasses existing ALS inhibitors in efficacy against Aspergillus fumigatus and other pathogenic fungi. Structural studies via cryo-electron microscopy reveal a unique covalent binding interaction between compound 1 and ALS, distinct from known inhibitors, and finally, we demonstrate that ALS is essential for virulence in a mouse model of invasive aspergillosis. These findings position ALS as a promising target for antifungal development and demonstrate the potential of resistance gene-guided genome mining for antifungal discovery.

|
Scooped by
mhryu@live.com
February 1, 1:02 PM
|
Glycosyltransferases (GTs) catalyze the formation of new glycosidic bonds and thus are vital for synthesizing nature's vast repertoire of glycans and glycoconjugates and for engineering glycan-related medicines and materials. However, obtaining detailed structural and functional insights for the >750,000 known GTs is limited by difficulties associated with their efficient recombinant expression. Members of the GT-C fold, in particular, pose the most significant expression challenges due to the integration and folding requirements of their multiple membrane-spanning regions. Here, we address this challenge by engineering water-soluble variants of an archetypal GT-C fold enzyme, namely the oligosaccharyltransferase PglB from Campylobacter jejuni (CjPglB), which possesses 13 hydrophobic transmembrane helices. To render CjPglB water-soluble, we leveraged two advanced protein engineering methods: one that is universal called SIMPLEx (solubilization of IMPs with high levels of expression) and the other that is custom tailored called WRAPs (water-soluble RFdiffused amphipathic proteins). Each approach was able to transform CjPglB into a water-soluble enzyme that could be readily expressed in the cytoplasm of E. coli cells at yields in the 3-6 mg/L range. Importantly, solubilization was achieved without the need for detergents and with retention of catalytic function. Collectively, our findings demonstrate that both SIMPLEx and WRAPs are promising platforms for advancing the molecular characterization of even the most structurally complex GTs, while also enabling broader GT-mediated glycosylation capabilities within synthetic glycobiology applications.
|

|
Scooped by
mhryu@live.com
Today, 7:19 PM
|
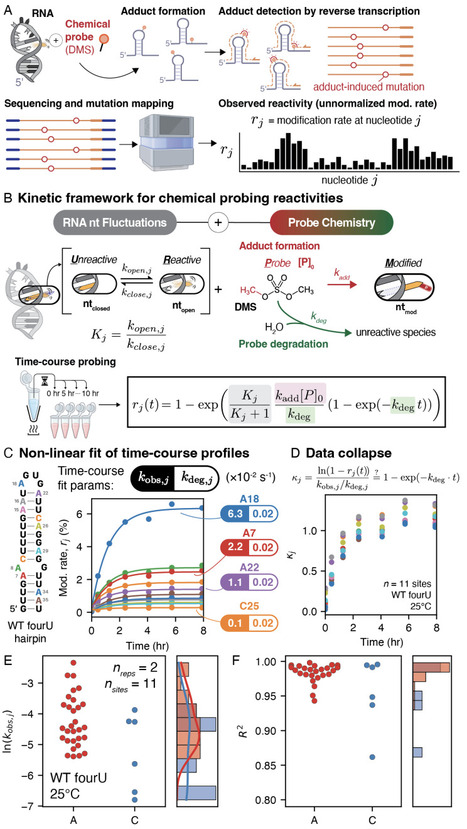
RNA function is governed by RNA folding, but strategies for measuring RNA folding thermodynamics are limited, and fundamental questions such as the energy of base pair opening remain debated. Here we introduce Probing-Resolved Inference of Molecular Energetics (PRIME) to extract nucleotide-resolution RNA structural energetics from scalable chemical probing experiments. Applying PRIME to diverse RNAs, we find that RNA base pairs and tertiary interactions dynamically open with free energies of 0.5-3 kcal/mol, revealing that RNA nucleotides ubiquitously sample open conformations at biologically accessible energies. PRIME further resolves energetic coupling across RNAs, providing an energetic understanding of RNA structural dynamics and long-range coordination in RNA folding. PRIME represents a widely accessible strategy for interrogating RNA thermodynamics, enabling mechanistic understanding and engineering of RNA biology.

|
Scooped by
mhryu@live.com
Today, 4:54 PM
|
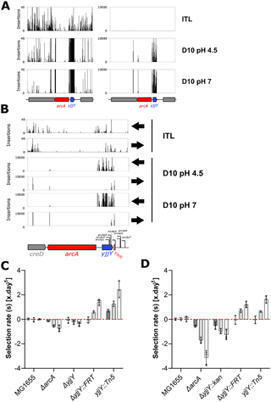
Long term laboratory evolution experiments are a powerful tool that are increasingly being used to study fundamental aspects of evolution and to identify genes that contribute to overall fitness under different conditions. However, even with automation, the time that they take to execute limits the extent to which evolution experiments can be used as part of a high throughput approach to understand the links between genotype and phenotype. Mutations that lead to genetic loss of function (LoF) are frequently selected for in evolution experiments. Thus, in principle these experiments could be done more rapidly by starting not with clonal isolates but with dense transposon libraries that will contain loss of function mutations in all non-essential genes. Here, we test this hypothesis by comparing the results of long term (5 month) evolution experiment, in which E. coli was grown with daily transfers in unbuffered LB starting at pH 4.5, with short term (5 and 10 day) experiments on a high-density transposon library in the same strain and under the same conditions. We show that there is a overlap in the genes and pathways identified using the two methods, as well as identifying other gene of interest whose LoF contributes to fitness. This approach has the potential to complement laboratory-based evolution, enabling rapid, higher throughput, testing of a wide range of parameters that may have an influence on evolutionary trajectories.

|
Scooped by
mhryu@live.com
Today, 4:33 PM
|
Synthetic biology is transforming how we understand and teach microbial energy metabolism. In a recent study (F. Li, B. Zhang, X. Long, H. Yu, et al., Nat Commun 16:2882, 2025, https://doi.org/10.1038/s41467-025-57497-z), the authors demonstrated a synthetic gene circuit that enables Shewanella oneidensis to produce and release phenazine-1-carboxylic acid, a redox-active metabolite that enhances extracellular electron transfer and electricity generation. This perspective highlights the significance of their work, focusing on how controlling the production of redox mediators provides new insights into microbial electron flow and bioelectronic design. Beyond its technological implications, this system also serves as a valuable educational case study for teaching principles of redox balance, gene regulation, and metabolic engineering. Viewing this advancement in the context of biology education underscores the potential of synthetic circuits to deepen our understanding of microbial metabolism and to promote interdisciplinary learning in microbiology, biotechnology, and engineering.

|
Scooped by
mhryu@live.com
Today, 4:21 PM
|
Antimicrobial resistance (AMR) poses a severe global health threat, with Acinetobacter baumannii among the critical AMR priorities highlighted by World Health Organization (WHO). This Gram-negative pathogen exhibits intrinsic resistance traits, exceptional environmental persistence, and high genomic plasticity, harboring resistance islands. To combat AMR through synthetic biology, this study characterizes a library of BioBrick parts to be adopted in A. baumannii engineering and develops a modular CRISPR interference (CRISPRi) platform. Key components were characterized, including two plasmid vectors, a library of inducible and constitutive promoters, and a CRISPRi-mediated repression system; for the latter, a testbed for biofilm-related genes implicated in the downregulation of antibiotic resistance is also provided. By enabling tunable transcriptional control through the characterized promoters and ensuring the ability to downregulate gene expression via CRISPRi, this synthetic biology toolkit lays the foundation for the rational design of genetic circuits to study and counteract AMR in A. baumannii. The modular platform here characterized provides a valuable resource for the iGEM community to advance functional genomic approaches against this alarming global health challenge.

|
Scooped by
mhryu@live.com
Today, 4:07 PM
|
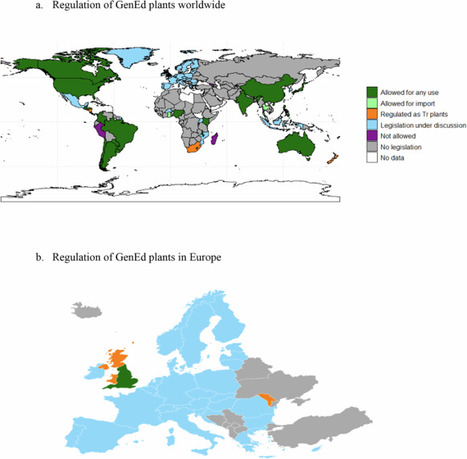
Precise breeding programs using new genome editing techniques have been developed to create plant varieties adapted to climate change. We studied the regulatory status of the 196 members of the United Nations (distinguishing the four UK countries) according to their legislation on transgenesis. We identified eight statuses for these techniques: ‘allowed for any use’ (24 countries, Argentina being the first in 2015), ‘allowed for import’ (1), ‘legislation under discussion’ (37), ‘not allowed except for food aid’ (0), ‘not allowed’ (3), ‘regulated as transgenics’ (7), ‘no legislation’ (114) and ‘no data available’ (10). We discussed the current situation in the European Union (EU) as many countries are awaiting its regulation. We also examined field trials carried out by six EU countries. We looked at authorised and commercialized gene-edited (GenEd) plants. Countries that have authorized transgenic (Tr) plants are 22.6% more likely to approve GenEd plants than those that have not.

|
Scooped by
mhryu@live.com
Today, 2:22 PM
|
CRISPR-Cas9 is natively present in the adaptive immune systems of a multitude of bacteria and has been adapted as an effective genome engineering tool. The Cas9 effector enzyme, which is composed of a single Cas9 protein and a single-guide RNA (sgRNA), identifies and cleaves double-stranded DNA targets through a series of conformational changes that require DNA distortion and unwinding. While most studies of Cas9 specificity have focused on the DNA sequence, the role of intrinsic DNA physical properties (“DNA shape”) in modulating Cas9 activity remains insufficiently defined. We previously showed that with a 16-nucleotide (-nt) truncated guide, the intrinsic DNA duplex dissociation energy at the PAM+(17–20) segment beyond the RNA–DNA hybrid tunes Cas9 cleavage rates of linear substrates. Here, we examined the impact of DNA supercoiling on Cas9 cleavage with the 16-nt truncated guide. Enzyme kinetic analysis revealed that PAM+(17–20) DNA sequences beyond the RNA/DNA hybrid preserve their effects on Cas9 cleavage in the supercoiled state. Furthermore, combining a novel asymmetric hairpin construct with a parallel-sequential kinetics model, rates for first-step nicking and second-step cleavage by Cas9 were obtained for both supercoiled and linear substrates. With both topologies, it was found that first-step nicking is clearly impacted by PAM+(17–20) DNA sequences, and the effects can be correlated with DNA unwinding, which dictates R-loop dynamics. This work expands our understanding of DNA target recognition by Cas9, and the methods developed, in particular those for analyzing the progression of Cas9-induced nicks, will aid in further in-depth mechanistic investigation.

|
Scooped by
mhryu@live.com
Today, 12:56 PM
|
Crops increasingly face overlapping stresses such as heat, drought, salinity, and pathogens that conventional breeding or genome editing rarely overcome in combination. To address this, we propose CRISPR-enabled horizontal gene transfer (CRISPR-HGT) as a programmable framework that recreates the evolutionary process by which plants historically acquired adaptive microbial genes. Microbial genes, refined under extreme environments, provide a naturally preadapted resource for multi-trait resilience. By integrating tools such as Cas12a, CasΦ, RNA-targeting, and dCas-based epigenome editors with AI-guided microbial gene discovery, CRISPR-HGT enables modular and inducible stress regulation. This approach shifts genome editing from allelic modification to evolution-guided design. We outline a conceptual pipeline spanning microbial gene mining to adaptive field deployment, highlighting the ecological, biosafety, and regulatory dimensions, from the European Union's cautious oversight to the UK's product-based framework. CRISPR-HGT thus introduces an evolution-informed paradigm for engineering crops that anticipate stress and sustain yield under climate uncertainty.

|
Scooped by
mhryu@live.com
Today, 12:38 PM
|
The growth of RNA sequencing (RNA-seq) data accompanied by the development of novel scalable data analytic methods has revealed a deep understanding of the composition of bacterial transcriptomes. This new, first-biological-principles understanding has enabled a novel characterization of the function of the transcriptional regulatory network. Here, we present a single-strain wild-type transcriptomic knowledgebase for the model strain E. coli MG1655. The associated transcriptomic compendium consists of 584 high-quality RNA-seq samples from wild-type E. coli MG1655 generated using a single protocol. These samples range over a wide condition space, including 45 carbon sources and 10 base media. Using independent component analysis, we decomposed the transcriptomic compendium to extract 115 independently modulated sets of genes (iModulons). We find that (i) iModulons explain 75% of variance in the dataset through knowledge enrichment; (ii) 67% of iModulons are associated with single/combined dominant regulators; (iii) iModulon activity profiles of samples can be utilized to elucidate patterns within the transcriptional regulatory network, such as differences in aerobicity; and (iv) the use of transcriptomic data derived from non-wild-type strains results in changes in iModulon gene membership, highlighting the malleability of the transcriptional regulatory network. Altogether, this knowledgebase serves as a resource for multi-scale knowledge mining for transcriptional regulation in E. coli MG1655.

|
Scooped by
mhryu@live.com
Today, 12:12 PM
|
Bacterial pathogens exploit host-derived nutrients to coordinate metabolism and virulence determinants to optimize fitness in vivo. In Pseudomonas aeruginosa, GlnK is a central regulator of nitrogen metabolism. It senses the intracellular nitrogen status by integrating 2-oxoglutarate (2-OG) and glutamine signals, which in turn triggers its uridylylation and conformational changes. This reversible post-translational modification modulates its interaction with target proteins, thereby precisely regulating carbon-nitrogen metabolic homeostasis and enabling adaptive nitrogen metabolism in response to host-derived nutrient cues. In this study, we found that glnK is upregulated during infection in a mouse pneumonia model. By growing bacteria in mouse bronchoalveolar lavage fluid (BALF), we demonstrated that the expression of glnK is activated by the NtrB-NtrC two-component regulatory system in response to the host nutrient environment. Mutation of glnK impairs bacterial virulence. Transcriptomic analysis revealed downregulation of the type III secretion system (T3SS) genes in the glnK mutant. Further studies revealed a role of GlnK in maintaining the homeostasis of the NtrB-NtrC system through a negative feedback mechanism, which is required for the expression of the T3SS genes. Collectively, these findings reveal a role of GlnK in interconnecting carbon–nitrogen balance and the T3SS in response to the host environment.

|
Scooped by
mhryu@live.com
Today, 10:42 AM
|
CRISPR–Cas enzymes must recognize a protospacer-adjacent motif (PAM) to edit a genomic site, greatly limiting the range of targetable sequences in a genome. Although engineering strategies to alter PAM specificity exist, they typically require labor-intensive, iterative experimentation. We introduce an evolution-informed deep learning model, Protein2PAM, to efficiently guide the design of Cas protein variants tailored to recognize specific PAMs. Trained on a dataset of over 45,000 CRISPR–Cas PAMs, Protein2PAM rapidly and accurately predicts PAM specificity directly from Cas proteins across type I, II and V CRISPR–Cas systems. Using in silico mutagenesis, the model identifies residues critical for PAM recognition in Cas9 without using structural information. We use Protein2PAM to computationally evolve Nme1Cas9, generating variants with broadened PAM recognition and up to a 50-fold increase in PAM cleavage rates compared to the wild type in vitro. Our machine learning approach allows Cas enzymes to target sequences that were previously inaccessible because of PAM constraints, potentially increasing target flexibility in personalized genome editing. The protospacer-adjacent motif specificity of several Cas enzymes is customized using protein language models.

|
Scooped by
mhryu@live.com
February 1, 1:33 PM
|
Genetic manipulation of core gut probiotics remains challenging due to endogenous cellular barriers and a scarcity of efficient molecular tools, limiting progress in live biotherapeutic development. Here, we characterized the native type II-C CRISPR-Cas system in Bifidobacterium longum subsp. longum GNB (B. longum GNB). Through integrated bioinformatic analysis and high-throughput protospacer adjacent motif (PAM) screening, we identified a novel 5′-NNRMAT-3′ (where R = A/G, M = A/C) motif recognized by its compact Cas9 nuclease (BLCas9). The stringent PAM dependency of BLCas9 was unequivocally confirmed by in vitro cleavage assays. Leveraging this endogenous mechanism, we developed a dual-plasmid editing platform for robust and multiplex genome engineering in the probiotic strain E. coli Nissle 1917. Application of this system notably enhanced extracellular γ-aminobutyric acid (GABA) production in EcN through targeted metabolic engineering. Our work provides the first molecular dissection of a type II-C system in Bifidobacterium longum and establishes a generalizable framework for the discovery and application of compact programmable nucleases, suggesting a viable strategy for modulating host physiology via the gut-brain axis.

|
Scooped by
mhryu@live.com
February 1, 1:08 PM
|
Cyclic dinucleotide (CDN) signaling molecules, such as cyclic di-GMP (c-di-GMP) and 3',3' cyclic GMP-AMP (cGAMP), are second messengers that play critical roles in phenotypic regulation, such as biofilm formation, host colonization, and bacterial virulence. Recently, hybrid promiscuous (Hypr) GGDEF proteins have been identified in certain bacteria to produce both cyclic dinucleotides. One such enzyme, Bd0367, from the predatory Bdellovibrio bacteriovorus, switches between synthesizing c-di-GMP and cGAMP to regulate the bacterial predation cycle and prey exit. However, the molecular mechanism controlling this switch remains unknown. Here, we introduce an RNA-based ratiometric, dual metabolite biosensor that enables simultaneous detection of c-di-GMP and cGAMP in live cells. This sensor integrates a Pepper-based biosensor for c-di-GMP detection and a Spinach2-based biosensor for cGAMP detection into a single transcript, producing distinct fluorescent outputs. In E. coli, the dual metabolite sensor reliably reported shifts in c-di-GMP/cGAMP production ratios from various CDN synthases, including Bd0367. Additionally, a histidine kinase was discovered as the probable regulatory partner of Bd0367. These findings demonstrate the sensor's capacity to assess relative CDN levels and to uncover complex signaling pathways. Together, this ratiometric dual metabolite biosensor provides a foundation for broader applications of fluorogenic RNA biosensors in dissecting bacterial signaling networks, microbial ecology, and host-pathogen interactions.
|
 Your new post is loading...
Your new post is loading...