Your new post is loading...

|
Scooped by
?
November 24, 11:56 PM
|
Core promoters comprise multiple elements whose interaction with RNA polymerase initiates transcription. Despite decades of research, substantial sequence and length variation of promoter elements has hindered efforts to elucidate their function and the evolutionary diversity of transcriptional regulation. Combining massively parallel assays, biophysical modeling, and functional validation, we systematically dissected the promoter architecture upstream of experimentally determined transcription start sites in 49 phylogenetically diverse bacterial genomes (GC content: 27.8%–72.1%). We identified a conserved 3-bp promoter element, termed “start,” which dictates transcription start site selection and enhances transcription. We uncovered a four-region organization within the variable spacer element, whose sequence composition modulates transcription by up to 600-fold. We showed that the discriminator element is conserved in Terrabacteria but diverse in Gracilicutes, the two major bacterial clades. High discriminator sequence diversity in Gracilicutes likely reflects diversifying evolution, enabling promoter-encoded regulation to orchestrate global gene expression in response to growth rate changes. Together, our findings reveal broad conservation of bacterial promoter organization while highlighting regulatory divergence of promoter elements and RNA polymerase between Terrabacteria and Gracilicutes. Sequence and functional similarities between bacterial promoter elements and their archaeal and eukaryotic counterparts further suggest a shared evolutionary origin of promoter architecture.

|
Scooped by
?
November 24, 11:41 PM
|
Bacterial flagella are known for facilitating motility to support nutrient acquisition and predator evasion, but can also serve as receptors for phages. Here, we characterize a dual role of flagella during infection of Yersinia enterocolitica by phage X1: functioning as a conduit for phage DNA entry and as a trigger for bacterial defense. X1 employs a “contraction-driven” mechanism, using its contractile tail to inject DNA into Yersinia flagella. Unlike other characterized flagellotropic phages that rely exclusively on flagella or less efficiently on secondary surface receptors, X1 can infect cells via lipopolysaccharides with even higher efficiency. Furthermore, certain Yersinia strains can detect X1 invasion and activate a flagellum-dependent toxin–antitoxin (Flag-TA) system, which requires the flagellar motor stator proteins MotAB to activate the abortive infection response. Our findings reveal a novel phage infection strategy and uncover a new bacterial defense that are both based on the flagellar machinery.

|
Scooped by
?
November 24, 11:28 PM
|
Communities of bacteria undergo population bottlenecks which are crucial to their population, ecological, and evolutionary dynamics. However, conventional amplicon sequencing cannot distinguish such demographic shifts between very closely related lineages. Here we describe BarTn7, an optimized method for bacterial lineage tracking through chromosomal integration of phenotypically neutral DNA barcodes with transposon Tn7. By combining conventional conjugative plasmids into a single vector and leveraging a parts-based strategy to optimize delivery to different recipients, BarTn7 increases barcoding efficiency and enables the systematic application of this tool to diverse bacteria. We tested BarTn7 in multiple bacterial species and confirmed a lack of lineage-specific growth effects. We then used BarTn7 to measure the colonization of Panicum virgatum roots under differing phosphate concentrations. Lineage tracking enabled discrimination between unique colonization events and the proliferation of existing bacteria during root colonization, the ratio of which differed between three plant-associated bacterial species. The strategy further allowed the measurement of phosphate-dependent ingress rates of the root endosphere by Paraburkholderia phytofirmans PsJN. We then demonstrated the effectiveness of BarTn7 to detect adaptive mutation(s) and facilitate identification of mutant lineages. Additionally, we demonstrated that BarTn7 is more accurate than conventional 16S rRNA gene sequencing at measuring community composition of a 5-member synthetic bacterial community (SynCom) and compares favorably to shotgun metagenomics. Our results illustrate the utility of BarTn7 as a simple, cost-efficient, and broadly applicable method of measuring bacterial population dynamics at sub-species resolution.

|
Scooped by
?
November 24, 11:19 PM
|
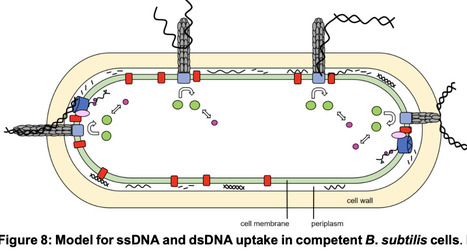
Natural competence describes the ability of several bacterial species to take up DNA from the environment for integration into the host chromosome, allowing the acquisition of novel genetic information. Uptake in many species operates via a dynamic pilus structure that pulls DNA through the cell wall into the periplasm by pilus retraction, followed by transport of one of the two DNA strands into the cytosol by an active membrane transporter. How double stranded DNA with a bending contour of hundreds of base pairs can fit through a narrow pore accommodating a pilus has been a puzzling question. We show that purified major and minor pilins from two Bacillus species, B. subtilis and Geobacillus thermodenitrificans, show efficient binding to double as well as to single stranded DNA with similar affinities. Consistent with uptake, ssDNA accumulates and moves within the periplasm before transport through the cell membrane, with similar dynamics as dsDNA. Thus, Bacillus competence pili are dsDNA as well as ssDNA uptake machines. The binding of pili to ssDNA regions within environmental DNA likely increases the efficiency of competent bacteria to pull extended DNA strands through the cell wall. Uptake of environmental ssDNA may also provide a beneficial source of nutrition for bacteria during stationary.

|
Scooped by
?
November 24, 7:27 PM
|
ArsR-based whole-cell biosensors offer sensitive colorimetric detection of arsenite [As(III)], yet their broad reactivity toward Group 15 metalloids—especially antimonite [Sb(III)]—limits field specificity. The recently identified ant operon from Comamonas testosteroni JL40 confers Sb(III)-selective resistance via the efflux ATPase AntA, the metallochaperone AntC, and the regulator AntR, providing genetic parts to suppress Sb(III) cross-talk. We systematically introduced antA, antC, and three antR homologs into an ArsR regulator coupled with a deoxyviolacein reporter chassis (pJ23119-K12). Co-expression of AntA and AntC under a moderate constitutive promoter (PceuR) shifted the Sb(III) limit of detection (LOD) from 0.073 µM to 0.586 µM, with a modest increase in the As(III) LOD to 0.018 µM. Subsequent integration of AntR1 not only maintained the As(III) LOD at 0.018 µM but also unexpectedly amplified the As(III) signal, extending the linear range to 36 nM–37.5 µM (R² = 0.991). It suggests that AntR1 may modulate the transcriptional circuitry via cross-regulation, warranting further mechanistic inquiry. The modified biosensor TOP10/pJ23119-antACR1 exhibited high selectivity for As(III) over divalent metals (Cd, Pb, Cu, Hg, Mn, Mg) and tolerated Sb(III) up to 1 µM. Performance was retained in 90% freshwater and 50% seawater matrices, enabling accurate quantification of 0–2.5 µM As(III) in deionized, tap, surface, and marine samples. By coupling Sb(III)-specific ant efflux/sequestration components with an ArsR-based sensing module, we developed a portable, low-cost biosensor that overcomes longstanding As(III)/Sb(III) cross-reactivity and performs robustly in complex environmental waters.

|
Scooped by
?
November 24, 6:43 PM
|
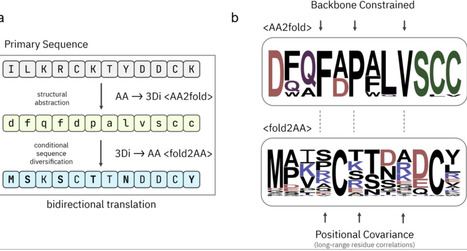
The accuracy of protein structure prediction models such as AlphaFold2 is tightly coupled to the depth and quality of multiple sequence alignments (MSAs), posing a persistent challenge for proteins with few or no identifiable homologs. We present GhostFold, a method for conjuring structure-constrained synthetic MSAs from a single amino acid sequence, bypassing the need for traditional homology searches. Leveraging the ProstT5 protein language model and the 3Di structural alphabet, GhostFold projects a query sequence into a tokenized structural representation and iteratively back-translates to generate an ensemble of diverse, fold-consistent sequences. These synthetic alignments (pseudoMSAs) encode emergent coevolutionary constraints that are sufficient for high-accuracy structure prediction of difficult targets such as orphan proteins and hypervariable antibody loops. GhostFold consistently matches or exceeds the performance of MSA-based and language model-based structure predictors while being computationally lightweight and independent of large sequence databases. Notably, we observe a decoupling of confidence metrics (e.g., pLDDT) from prediction accuracy when using pseudoMSAs, suggesting that AlphaFold2’s internal confidence calibration is strongly influenced by the statistical properties of natural sequence alignments. These results establish that structure-guided synthetic MSAs can functionally substitute for evolutionary data, offering a scalable and generalizable solution to one of the central limitations in computational structural biology. GhostFold represents a shift from passive data mining to intelligent sequence synthesis, redefining how structural priors are encoded in deep learning-based protein folding.

|
Scooped by
?
November 24, 4:49 PM
|
The diverse regulatory functions, protein production capacity, and stability of natural and synthetic RNAs are closely tied to their ability to fold into intricate structures. Determining RNA structure is thus fundamental to RNA biology and bioengineering. Among existing approaches to structure determination, computational secondary structure prediction offers a rapid and low-cost strategy and is thus widely used, especially when seeking to identify functional RNA elements in large transcriptomes or screen massive libraries of novel designs. While traditional approaches rely on detailed measurements of folding energetics and/or probabilistic modeling of structural data, recent years have witnessed a surge in deep learning methods, inspired by their tremendous success in protein structure prediction. However, the limited diversity and volume of known RNA structures can impede their ability to accurately predict structures markedly different from the ones they have seen. This is known as the generalization gap and currently poses a major barrier to progress in the field. In this Perspective article, we gauge method generalizability using a new benchmark dataset of structured RNAs we curated from the Protein Data Bank. We also discuss the emergence of deep learning methods for predicting structure probing data and use a new dataset to underscore generalization challenges unique to this domain along with directions for future improvement. Expanding beyond improving predictive accuracy, we review how advances in deep learning have recently enabled scalable and accessible optimization of traditional structure prediction methods and their seamless integration with modern neural networks.

|
Scooped by
?
November 24, 4:33 PM
|
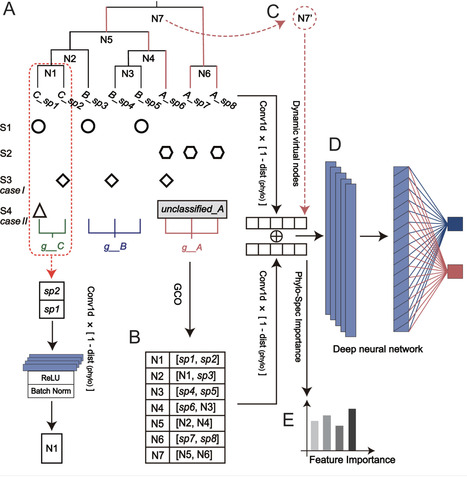
The human microbiome is crucial for health regulation and disease progression, presenting a valuable opportunity for health state classification. Traditional microbiome-based classification relies on pre-trained machine learning (ML) or deep learning (DL) models, which typically focus on microbial distribution patterns, neglecting the underlying relationships between microbes. As a result, model performance can be significantly affected by data sparsity, misclassified features, or incomplete microbial profiles. To overcome these challenges, we introduce Phylo-Spec, a phylogeny-driven deep learning algorithm that integrates multi-aspect microbial information for improved status recognition. Phylo-Spec fuses convolutional features of microbes within a phylogenetic hierarchy via a bottom-up iteration and significantly alleviates the challenges due to sparse data and inaccurate profiling. Additionally, the model dynamically assigns unclassified species to virtual nodes on the phylogenetic tree based on higher-level taxonomy, minimizing interferences from unclassified species. Phylo-Spec also captures the feature importance via an information gain-based mechanism through the phylogenetic structure propagation, enhancing the interpretability of classification decisions. Phylo-Spec demonstrated superior efficacy in microbiome status classification across two in silico synthetic data sets that simulate the aforementioned cases, outperforming existing ML and DL methods. Validation with real-world metagenomic and amplicon data further confirmed the model’s performance in multiple status classification, establishing a powerful framework for microbiome-based health state identification and microbe-disease association. The source code is available at https://github.com/qdu-bioinfo/Phylo-Spec.

|
Scooped by
?
November 24, 4:12 PM
|
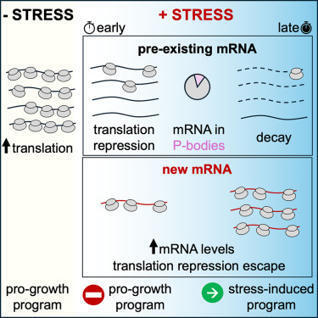
When cells encounter stress, they rapidly mount an adaptive response by switching from pro-growth to stress-responsive gene expression programs. How cells selectively silence pre-existing, pro-growth transcripts yet efficiently translate transcriptionally induced stress mRNA and whether these transcriptional and post-transcriptional responses are coordinated are poorly understood. Here, we show that following acute glucose withdrawal in S. cerevisiae, pre-existing mRNAs are not first degraded to halt protein synthesis, nor are they sequestered away in P-bodies. Rather, their translation is quickly repressed through a sequence-independent mechanism that differentiates between mRNAs produced before and after stress, followed by their decay. Transcriptional induction of endogenous transcripts and reporter mRNAs during stress is sufficient to escape translational repression, while induction prior to stress leads to repression. Our results reveal a timing-controlled coordination of the transcriptional and translational responses in the nucleus and cytoplasm, ensuring a rapid and wide-scale reprogramming of gene expression following environmental stress.

|
Scooped by
?
November 24, 3:57 PM
|
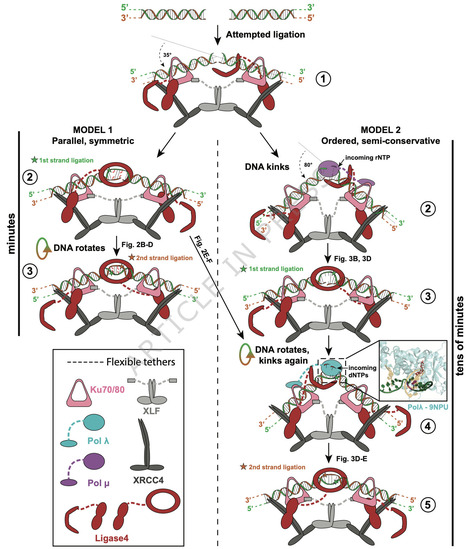
Nonhomologous end-joining repairs chromosomal double strand breaks, but it is unknown whether both strands are repaired by this pathway, and if one strand break’s repair path impacts the other. Here, we show that nonhomologous end-joining employs both of two a priori possible strategies. Strand breaks that can be directly ligated are joined near-simultaneously, with no effect of one strand break’s repair path on the other. More complex end structures require obligatorily ordered repair. The first strand to be repaired is used as template for repair of the opposite/second strand break, with the latter repair reaction occurring fastest when also coupled to nonhomologous end-joining. Enforced asymmetry in repair of each strand break can extend to the gap-filling polymerase employed, and whether the polymerases incorporate RNA or DNA. Our results resolve questions about pathway mechanism and identify a requirement for flexibility of the nonhomologous end-joining machinery for efficient repair of both strand breaks within diverse cellular double strand breaks. Breakage of both chromosomal DNA strands creates unique problems for DNA repair. Here, Luthman et al. show that for some broken ends, the two strand breaks are repaired in parallel, while for other ends one strand must be repaired before the other.

|
Scooped by
?
November 24, 3:36 PM
|
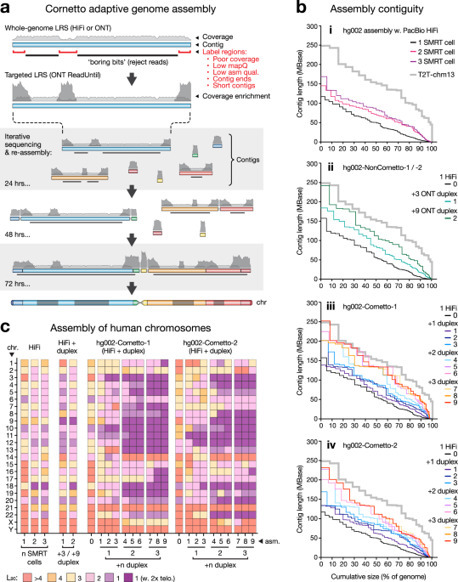
Advances in long-read sequencing (LRS) and assembly algorithms have made it possible to create highly complete genome assemblies for humans, animals and plants. However, ongoing development is needed to improve accessibility, affordability, and assembly quality and completeness. ‘Cornetto’ is a new strategy in which we use programmable selective nanopore sequencing to focus LRS data production onto the unsolved regions of a nascent assembly. This improves assembly quality and streamlines the process, both for humans and non-human vertebrates. Cornetto enables us to generate highly complete diploid human genome assemblies using only nanopore LRS data, surpassing the quality of previous efforts at a fraction of the cost. Cornetto enables genome assembly from challenging sample types like human saliva. Finally, we obtain accurate assemblies for clinically-relevant repetitive loci at the extremes of the genome, demonstrating valid approaches for genetic diagnosis in facioscapulohumeral muscular dystrophy (FSHD) and MUC1-autosomal dominant tubulointerstitial kidney disease (MUC1-ADTKD). Long-read sequencing enables high-quality genome assemblies, but challenges remain. Here, the authors introduce Cornetto, a method that improves assembly quality, enables genome sequencing from saliva, and accurately resolves medically-relevant repetitive genes.

|
Scooped by
?
November 24, 1:52 PM
|
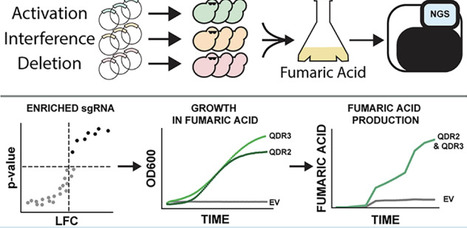
Organic acids such as fumaric acid are widely used in the food and beverage industry as acidulants and preservatives, while also serving as versatile precursors for industrially relevant compounds. Fumaric acid is still predominantly produced through petroleum-derived processes. To enhance production efficiency and diversify supply, we are engineering Kluyveromyces marxianus as a biosynthetic platform from renewable feedstocks. In previous work, we have established K. marxianus Y-1190 as a host for lactose valorization based on its high growth rate on lactose and its tolerance for acid conditions. Here, we establish a trifunctional genome-wide library for K. marxianus using CRISPR activation, interference, and deletion to allow identification of gene expression perturbations that enhance tolerance to fumaric acid. We determined that deletion of ATP7, encoding a subunit of the mitochondrial F1F0 ATP synthase, and overexpression of QDR2 and QDR3, two previously uncharacterized members of the 12-spanner H+ antiporter (DHA1) family in K. marxianus, can enhance fumaric acid tolerance. We also found that integrated overexpression of both QDR2 and QDR3 in a ΔFUM1 background strain improved titers of fumaric acid production from 0.26 to 2.16 g L–1. Together, these results highlight roles for membrane transport and mitochondrial function in enabling fumaric acid tolerance and production in K. marxianus.

|
Scooped by
?
November 24, 1:40 PM
|
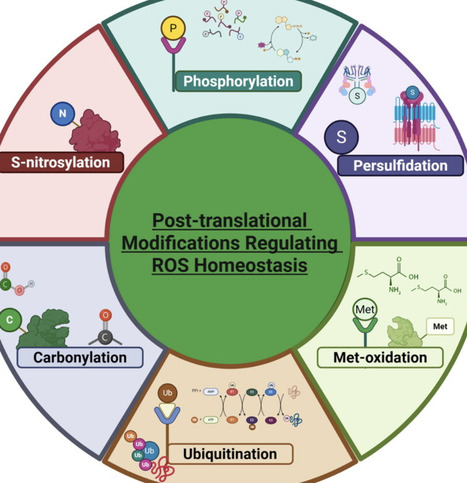
Reactive oxygen species (ROS) play crucial roles in many plant biological processes. ROS have emerged as major signaling molecules mediating various regulatory reactions in response to environmental stimuli. This signaling is mediated by a highly regulated process of ROS accumulation at specific cellular compartments. Therefore, this review focuses on the intricate ROS signaling in plant defense and strategic virulence effectors from pathogens hijacking ROS homeostasis. In addition, the ROS-mediated regulation of plant processes acts through post-translational modifications (PTMs) is discussed. We also provide a valuable roadmap for translating ROS research into climate-resilient cultivars by exploring the manipulation of pathogen effectors, ROS cascade genes through modern biotechnological approaches, and post-translational modifications against various environmental stressors. This framework can advance research in plant stress biology and agricultural practices.
|

|
Scooped by
?
November 24, 11:44 PM
|
CRISPR activation is a powerful tool to upregulate a vast array of genes in many different contexts. However, there are few dynamic CRISPR transcriptional programs, which limit its usage in the creation of living biosensors, self-regulating microbial factories, or conditional therapeutics. Here, we address this limitation by embedding a molecular switch directly into a guide RNA to create a combined sensor–actuator called a metabolite-responsive scaffold RNA (MR-scRNA). We demonstrate the regulatory potential for MR-scRNAs by conditionally activating genes in three different kingdoms of life. We create MR-scRNAs responsive to two distinct metabolites, theophylline and tryptophan, by swapping the molecular switch used. MR-scRNAs respond quickly in a dose-dependent manner specifically to their target metabolite and enhance biochemical production when used as a dynamic regulator of pathway enzyme expression. The broad functionality and ease of design of the MR-scRNAs offer a promising tool for dynamic cellular regulation.

|
Scooped by
?
November 24, 11:37 PM
|
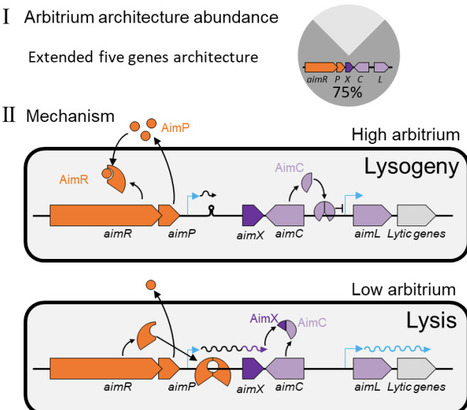
Many temperate Bacillus phages utilize the arbitrium signaling system to control lysis/lysogeny decisions. While the function of the arbitrium signal AimP and its receptor AimR are well known, it is unclear how they control lysis in most arbitrium systems. Here, we show that a large majority of arbitrium systems are embedded in an extended module with three additional components; A small antirepressor protein (AimX), the phage repressor (AimC) and an adjacent cro-like protein (AimL). AimR-dependent activation of AimX is necessary for lysis both during infection and lytic induction. Molecular analysis suggests that AimX directly binds AimC and prevents its oligomerization and binding to its regulated aimL promoter. Our work therefore uncovers the main mechanism by which arbitrium systems regulate lysis and point to the central role of small proteins in phage decision making.

|
Scooped by
?
November 24, 11:24 PM
|
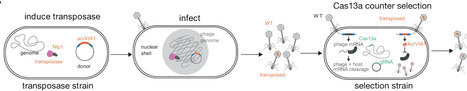
Bacteriophages have genomes that span a wide size range, are densely packed with coding sequences, and frequently encode genes of unknown function. Classical forward genetics has defined essential genes for phage replication in a few model systems but remains laborious and non-scalable. Unbiased functional genomics approaches are therefore needed for phages, particularly for large lytic phages. Here, we develop a phage transposon sequencing (Tn-Seq) platform that uses the mariner transposase to insert an anti-CRISPR selectable marker into phage genomes. CRISPR-Cas13a-based enrichment of transposed phages followed by pooled sequencing identifies both fitness-conferring and dispensable genes. Using the Pseudomonas aeruginosa-infecting nucleus-forming jumbo phage ΦKZ (280,334 bp; 371 predicted genes) as a model, we show that ~110 genes are fitness-conferring via phage TnSeq. These include conserved essential genes involved in phage nucleus formation, protein trafficking, transcription, DNA replication, and virion assembly. We also isolate hundreds of individual phages with insertions in non-essential genes and reveal conditionally essential genes that are specifically required in clinical isolates, at environmental temperature, or in the presence of a defensive nuclease. Phage TnSeq is a facile, scalable technology that can define essential phage genes and generate knockouts in all non-essential genes in a single experiment, enabling conditional genetic screens in phages and providing a broadly applicable resource for phage functional genomics.

|
Scooped by
?
November 24, 11:04 PM
|
Advances in CRISPR technologies have led to the use of diverse CRISPR-associated (Cas) proteins in diagnostic applications. Herein, we present a CRISPR/Cas12a2-based amplification-free RNA detection method that exhibits sub-attomolar sensitivity and substrate versatility. Cas12a2, a recently characterized RNA-guided nuclease, uniquely integrates bimolecular recognition through CRISPR RNA (crRNA)-target complementarity and protospacer flanking sequence identification, enabling highly specific trans-cleavage of single-stranded DNA, double-stranded DNA, and RNA. We have optimized key biochemical parameters, including pH, ionic strength, and temperature, to enhance the catalytic efficiency of Cas12a2. Based on the optimal activity conditions of Cas12a2, we have achieved ultra-sensitive viral RNA detection with a limit of detection of 46.7 aM through the strategic design and cooperative activation of crRNAs targeting conserved regions of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome. The diagnostic accuracy of the Cas12a2-based assay has been demonstrated for 26 SARS-CoV-2 variants, and it has further resulted in the definitive diagnosis of 317 clinical samples. This work establishes Cas12a2 as a promising molecular diagnostic tool that provides an amplification-free, rapid, and versatile solution for RNA detection. The adaptability and simplicity of the platform render it particularly well suited for point-of-care applications, paving the way for next-generation CRISPR diagnostics.

|
Scooped by
?
November 24, 7:05 PM
|
Kluyveromyces marxianus has high biotechnological potential, particularly because it can utilize a wide range of substrates and grow faster than other yeasts. It has also attracted interest as a prospective probiotic; however, this application is still not well researched. Two K. marxianus strains with promising biotechnological properties and the proven ability to efficiently produce 2-phenylethanol (2-PE) and ethanol were examined in this study to evaluate their potential as probiotics. Yeasts WUT216 and WUT240 exhibited similar survival rates in a simulated human digestive system as the reference K. marxianus strain B0399 (DiarYeast®, LongLife, Milano, Italia). Additionally, the WUT yeasts revealed a high degree of adhesion to cancer and normal cell lines in vitro. Importantly, the WUT strains did not exhibit undesirable characteristics, such as mucolytic and hemolytic activities, and toxic effects on the Caco-2 cell lines in vitro and on Galleria mellonella larvae in vivo. Furthermore, K. marxianus strains displayed attractive probiotic properties, particularly high antioxidant potential, antifungal activity, and significant β-glucan content. This study also showed that the WUT yeasts are sensitive to the commercial antifungals, fluconazole, amphotericin B, and geneticin G418. Overall, the WUT240 and WUT216 exhibit similar probiotic properties to those of B0399. Thus, the WUT yeasts are not only promising producers of 2-PE and ethanol but also attractive probiotic candidates. Additionally, discrepancies in literature reports regarding the analyses conducted were noted, particularly in adhesion and autoaggregation tests.

|
Scooped by
?
November 24, 5:19 PM
|
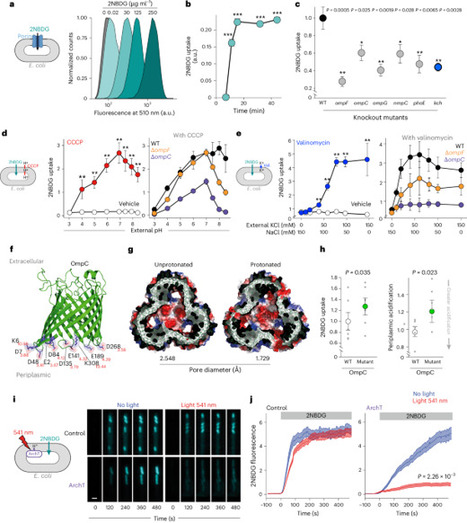
Porins mediate the passage of hydrophilic nutrients and antibiotics across the outer membrane but might contribute to proton leak from the periplasm, suggesting that their conductance could be regulated. Here we show, using single-cell imaging, that porin permeability in Escherichia coli is controlled by changes in periplasmic H+ and K+ concentration. Conductance through porins increases with low periplasmic H+ caused by starvation, promoting nutrient uptake, and decreases with periplasmic acidification during growth in lipid media, limiting proton loss. High metabolic activity during growth in glucose media, however, activates the inner membrane voltage-gated potassium channel, Kch, increasing periplasmic potassium and enhancing porin permeability to dissipate reactive oxygen species. This metabolic control of porin permeability explains the observed increase in ciprofloxacin resistance of bacteria catabolizing lipids and clarifies the impact of mutations in central metabolism genes on drug resistance, identifying Kch as a therapeutic target to improve bacterial killing by antibiotics. The permeability of bacterial porins is dynamically regulated by periplasmic proton and potassium concentrations, altering antibiotic resistance.

|
Scooped by
?
November 24, 4:39 PM
|
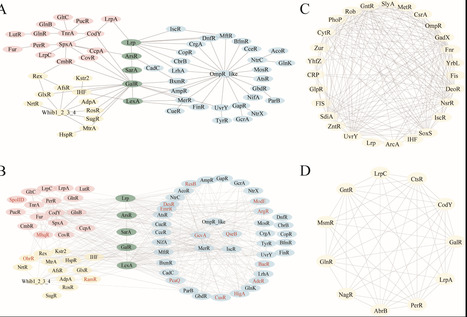
Global regulators (GRs) are key transcription factors that orchestrate the expression of multiple genes, playing essential roles in stress responses, virulence, secondary metabolism, and antibiotic resistance—traits that make them powerful tools for synthetic biology applications. However, conventional approaches often fail to detect remote homologs and novel GR types, limiting our understanding of their regulatory diversity and evolutionary dynamics across prokaryotes. Here, we present a large-scale, protein language model-driven framework to systematically chart the global regulatory landscape across 14,800 bacterial and archaeal type strain genomes—the most taxonomically diverse prokaryotic data set analyzed to date. Using a deep learning-based GR identification model trained on 74,872 curated GR sequences, we systematically identified over 270,000 GR-like proteins, including 173,256 homologs of 214 experimentally validated GR types, 52 putative GR types, and 76,113 proteins of unknown function. This model demonstrated high sensitivity and generalization capability, enabling the discovery of remote homologs and cryptic regulators beyond the reach of similarity- or domain-based methods. This expanded GR catalog revealed lineage-specific distribution patterns, functionally diverse regulons with both conserved and niche-specific targets, and hierarchical cross-regulatory networks with shared and phylum-enriched hubs. By unveiling the hidden diversity and evolutionary structure of prokaryotic global regulators, this landscape of GRs provides foundational insights into microbial gene regulation and offers a powerful toolkit for the rational design of tunable, modular, and orthogonal genetic circuits in synthetic biology.

|
Scooped by
?
November 24, 4:24 PM
|
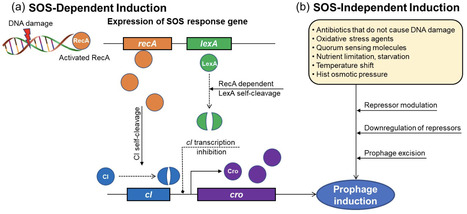
Bacteriophages (phages), the dominant prokaryotic viruses that specifically target bacteria in the human gut microbiome, play a crucial role in maintaining intestinal balance, regulating bacterial populations, and preserving microbial diversity within the gut microbiota. While prophages can enhance bacterial virulence and antibiotic resistance, potentially posing health risks, they also provide beneficial functions, including enhancing host fitness, promoting immune modulation, and contributing to ecosystem resilience, which supports intestinal homeostasis. Human gut microbiota is essential for various physiological functions, including digestion, vitamin synthesis, immune modulation, and protection against pathogens. Dysbiosis, or microbial imbalance, is associated with various disorders such as inflammatory bowel disease, obesity, diabetes, and mental health disorders. Consequently, prophages are important considerations for developing therapies to prevent intestinal diseases. Recently, there has been significant interest in prophage induction in the gut due to its functional impacts on microbial dynamics, gut health, and disease modulation. Prophage induction can be regulated by diet, antibiotics, metabolites, gut health, lifestyle, and intestinal environments. However, compared with lytic phages, prophages remain underexplored, leaving gaps in our understanding of their functions within the gut. Therefore, further research is needed to fully elucidate the complex interactions between phages, prophages, and the gut microbiota, and their effects on health and disease. This knowledge could inform the development of phage-based therapies and improve therapeutic strategies for gut health.

|
Scooped by
?
November 24, 4:05 PM
|
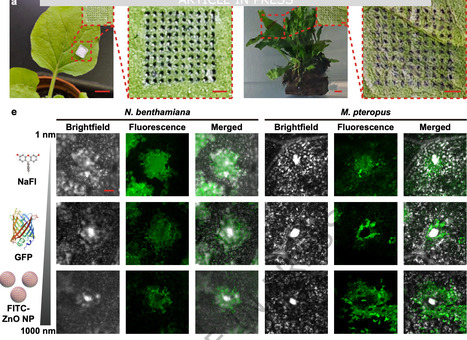
Efficient cargo delivery is essential for plant trait engineering, yet existing methods are often species-specific and ineffective across diverse habitats. Here, we develop core-shell microneedles for targeted delivery of biomolecular cargoes and active microorganisms into both terrestrial and aquatic plants. The microneedle architecture is rationally engineered to resist water exposure and release cargo upon contact with plant interstitial fluid, enabling controlled delivery into tissues and cells. We demonstrate that these core-shell microneedles can efficiently transport diverse cargoes, from nanoscale biomolecules such as functional nucleic acids, proteins, and plant hormones to microscale bioactive Agrobacterium, leading to strong protein expression and enhanced plant growth. Underwater delivery of salt-tolerance genes into submerged freshwater plants further demonstrates the platform’s utility for engineering stress resilience in challenging environments. By facilitating the cellular uptake of diverse cargoes into intact plants across different habitats, this amphibious microneedle strategy offers a versatile cargo delivery tool to advance plant biotechnology and environmental applications. An efficient cargo delivery tool is essential for plant trait engineering. Here, the authors repot a core-shell microneedles for targeted delivery of nucleic acids, proteins, phytohormones, and Agrobacterium to both terrestrial and aquatic plants.

|
Scooped by
?
November 24, 3:46 PM
|
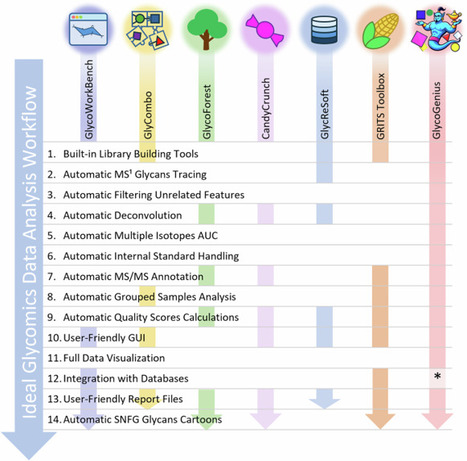
Mass spectrometry is recognized as the gold standard for glycan analysis, yet the complexity of the generated data hampers progress in glycobiology, as existing tools lack full automation, requiring extensive manual effort. We introduce GlycoGenius, an open-source program offering an automated workflow for glycomics data analysis, featuring an intuitive graphical interface. With algorithms tailored to reduce manual workload, it allows for data visualization and automatically constructs search spaces, identifies, scores, and quantifies glycans, filters results, and annotates fragment spectra of N- and O-glycans, glycosaminoglycans and more. It seamlessly guides researchers of all expertise levels from raw data to publication-ready figures. Our findings demonstrate that GlycoGenius achieves results comparable to manual analysis or competing tools, identifying more glycans, including novel ones, while significantly reducing processing time. This groundbreaking tool represents a significant advancement in the study of glycoconjugates, empowering researchers to focus on insights rather than data processing. Researchers present GlycoGenius, an open-source tool that automates complex glycomics data analysis. It streamlines workflows, identifies known and previously unreported glycans, and enables faster, more accessible insights into glycobiology.

|
Scooped by
?
November 24, 2:43 PM
|
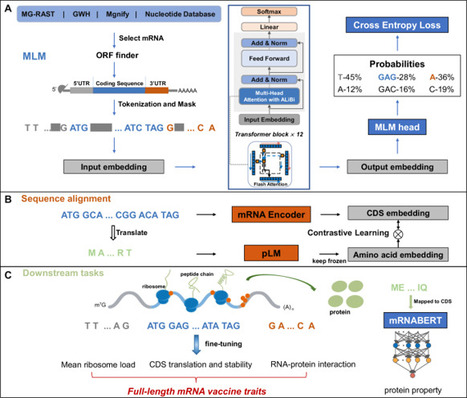
Designing effective mRNA sequences for therapeutics remains a formidable challenge. Inspired by successes in protein design, language models (LMs) are now being applied to RNA, but progress is often impeded by the lack of comprehensive training data. Existing models are frequently limited to UTR or CDS regions, restricting their application for complete mRNA sequences. We introduce mRNABERT, a robust, all-in-one mRNA designer pre-trained on the largest available mRNA dataset. To enhance performance, we propose a dual tokenization scheme with a cross-modality contrastive learning framework to integrate semantic information from protein sequences. On a comprehensive benchmark, mRNABERT demonstrates state-of-the-art performance, outperforming previous models in the majority of tasks for 5’ UTR and CDS design, RNA-binding protein (RBP) site prediction, and full-length mRNA property prediction. It also surpasses large protein models in several related tasks. In conclusion, mRNABERT’s superior performance across these diverse tasks signifies a substantial leap forward in mRNA research and therapeutic development. Designing complete mRNA sequences for new vaccines and therapies is a complex challenge. Here, the authors develop mRNABERT, a foundational AI model that designs entire mRNA sequences and demonstrates superior performance across comprehensive benchmarks.

|
Scooped by
?
November 24, 1:43 PM
|
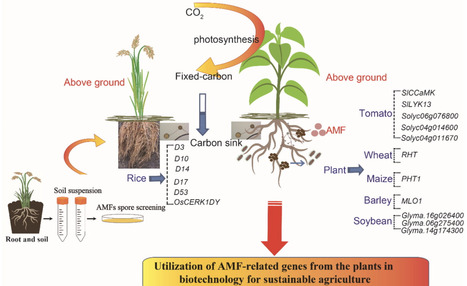
In recent years, interest in the role of nutrient cycling in sustainable agriculture has significantly increased. The potential of arbuscular mycorrhizal fungi (AMF) in nutrient cycling and plant growth improvement has long been recognized. However, there have been only a few studies on the identification and exploration of AM symbiosis-related plant genes for sustainable agriculture. We have developed a new constructive model for using host plant-derived AM symbiosis-related genes to improve breeding and AMF utilization for sustainable agriculture, particularly in the context of climate change. This model include: 1) the discovery of AM symbiosis-related genes in crop wild-relatives for molecular breeding and 2) the screening and propagation of AMFs that can help improve water-use efficiency and nutrient-use efficiency by crops, thereby reducing chemical fertilizer use in agricultural production. The first approach uniquely facilitates the identification of host plant-derived AM symbiosis-related genes, such as CHITIN ELICITOR RECEPTOR KINASE 1 (OsCERK1) from Dongxiang (DY) wild rice (Oryza rufipogon) (OsCERK1DY), MILDEW RESISTANCE LOCUS 1 (MLO1) from wild barley (Hordeum spontaneum), and WRKY60 from wild soybean (Glycine soja), for breeding purposes. The second one involves identifying soil-borne AMF species, such as Rhizophagus intraradices and Glomus mosseae for practical applications in the field. This suggestive model presents an emerging biotechnological potential for engineering climate-resilient crops.
|
 Your new post is loading...
Your new post is loading...