 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:24 AM
|
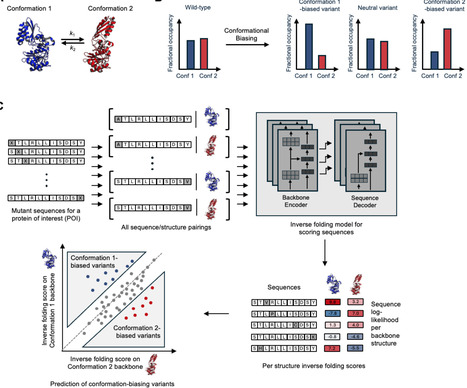
Conformational biasing (CB) is a rapid and streamlined computational method that uses contrastive scoring by inverse folding models to predict protein variants biased toward desired conformational states. We successfully validated CB across seven diverse datasets, identifying variants of K-Ras, SARS-CoV-2 spike, β2 adrenergic receptor, and Src kinase with improved conformation-specific functions, such as enhanced binding or enzymatic activity. Applying CB to the enzyme lipoic acid ligase (LplA), we uncovered a previously unknown mechanism controlling its promiscuous activity. Variants biased toward an “open” conformation state became more promiscuous, whereas “closed”-biased variants were more selective, enhancing LplA’s utility for site-specific protein labeling with fluorophores in living cells. The speed and simplicity of CB make it a versatile tool for engineering protein dynamics with broad applications in basic research, biotechnology, and medicine.

|
Scooped by
mhryu@live.com
Today, 1:08 AM
|
The dark-operative protochlorophyllide oxidoreductase (DPOR) catalyzes the light-independent reduction of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), a key step in photosynthetic pigment biosynthesis. Structurally and mechanistically related to nitrogenase, DPOR consists of a reductase (BchL) and a catalytic component (BchNB) homologous to the reductase (NifH) and catalytic component (NifDK) of Mo-nitrogenase. Structural alignment of Rhodobacter capsulatus (Rc) BchNB with Azotobacter vinelandii (Av) NifDK and the cofactor maturase NifEN reveals a conserved α2β2 architecture and a shared cofactor-insertion path linking their respective prosthetic-like group/cofactors (Pchlide, M-cluster, L-cluster), suggesting the possibility of generating chimeric proteins with novel reactivities. Herein, Pchlide-free RcBchNB (RcBchNBapo) is reconstituted with the L-cluster extracted from AvNifEN to yield a hybrid protein (RcBchNBL) capable of reducing N2 and C1 substrates (CN−, CO) to NH3 and hydrocarbons, respectively, in the presence of a strong reductant (EuII-DTPA). In contrast, reconstituting Pchlide-bound RcBchNB with the L-cluster yields minimal activity, indicating that Pchlide and the L-cluster compete for a common binding site, as supported by Boltz-2 modeling. These findings support the hypothesis of an intertwined evolution of photosynthetic and nitrogen-fixing enzymes and outline a framework for engineering chimeric metalloenzymes that couple light capture with nitrogenase-like catalysis in the future.

|
Scooped by
mhryu@live.com
January 11, 12:18 PM
|
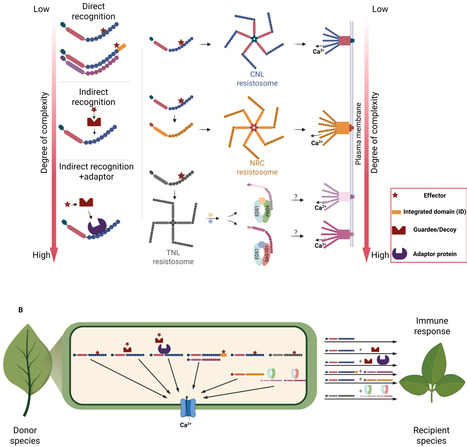
Plants employ cell-surface and intracellular immune receptors to perceive pathogens and activate defense responses. Recent advances in mechanistic understanding of how cell-surface and intracellular immune receptors convert recognition of molecular patterns or effectors into defense activation, combined with the knowledge of receptor repertoire variation both within and between species, allow transfer of immune receptors between species to increase the spectrum of recognition specificities. Here, we summarize recent progress in the functional transfer of immune receptors within and between plant families. We also discuss challenges that limit the transferability of intracellular immune receptors, including the requirement of additional host factors or downstream components and their incompatibility between donor and recipient species. Finally, we provide an overview of future perspectives for bioengineering disease-resistant crops through immune receptor transfer.

|
Scooped by
mhryu@live.com
January 11, 12:08 PM
|
The human gut microbiota, particularly the intestinal microbiota, shapes host physiology, disease risk, and therapeutic outcomes through complex metabolic and enzymatic activities. Recent advances in molecular omics, metabolomics, enzyme bioinformatics, and artificial intelligence (AI) have created unprecedented opportunities to elucidate its therapeutic roles to further enable precision microbiome medicine for personalized prevention, diagnosis, and treatment. In this review, we highlight emerging applications that leverage molecular omics and metabolomics technologies to dissect gut microbial functions, along with developments in enzyme bioinformatics and AI tools that reveal gut microbial species, enzymes, and metabolic pathways impacting human health. Finally, we discuss perspectives on data standardization, functional annotation, and interpretability, and how emerging tools are accelerating translational microbiome research.

|
Scooped by
mhryu@live.com
January 11, 11:52 AM
|
Lignin, a complex natural aromatic polymer, poses significant challenges to its efficient degradation, hindering the utilization of biomass for many industrial applications. Bacterial degradation of lignin may offer a promising solution to this challenge. This project aimed at elucidating the function of secreted oxidative enzymes from Pseudomonas putida involved in degradation and utilization of lignin and lignin-derived compounds. Using CRISPR-Cas9 and CRISPR-Cas3 systems, the putative lignin-degrading versatile peroxidase gene (VP; PP_1686, originally annotated as glutathione peroxidase GPx) and dye-decolorizing peroxidase gene (PP_3248) were individually knocked out from P. putida KT2440. The ∆PP_1686 mutant exhibited impaired growth and utilization of lignin-derived compounds. This correlated with reduced expression of p-hydroxybenzoate hydroxylase pobA and of DNA repair modules, alongside compensatory upregulation of energy and redox supply pathways. This work expands our knowledge on bacterial glutathione peroxidase by presenting a role beyond ROS scavenging. This work revealed the importance of P. putida VP/GPx in maintaining redox balance while supporting lignin-derived aromatic metabolism, offering new targets for future investigation into stress–metabolism crosstalk and lignin valorization strategies.

|
Scooped by
mhryu@live.com
January 11, 11:44 AM
|
RNA detection applications can be augmented if a sensed RNA can be directly functionally transduced. However, there is no generalizable approach that allows an RNA trigger itself to directly activate diverse non-coding RNA effectors. Here, we report engineering of a programmable, RNA trigger-activated, dual-site self-cleaving ribozyme with modular sensing domain and cleavage product. This platform, UNlocked by Activating RNA (UNBAR), is entirely encoded within one RNA strand. The ribozyme can be designed to be almost completely inactive in absence of trigger, and to exhibit single-nucleotide trigger specificity. UNBAR ribozymes carry out cell-free sensing and protein-free amplification of microRNA and viral RNA sequences, and trigger-dependent release of ncRNA effectors sgRNA, shRNA and aptamer. We demonstrate RNA detection and functional transduction by a cleaved aptamer, whose fluorescence can be directly read out as a function of trigger RNA. We further engineer the ribozyme for function in cells, and demonstrate trigger-dependent regulation of CRISPR-Cas9 editing by sgRNA-embedded ribozymes in zebrafish embryos and human cells. UNBAR is a first-in-class modality with potential to be developed into a versatile platform for synthetic biology, diagnostics and gene regulation. To enable a sensed RNA to activate diverse RNA effectors, the authors engineer a programmable dual-site ribozyme that, upon RNA trigger binding, self-cleaves to release an embedded RNA. It enables trigger-dependent release of diverse ncRNAs and controls CRISPR-Cas9 editing in zebrafish and human cells.

|
Scooped by
mhryu@live.com
January 10, 4:46 PM
|
The pathogenic bacterium Vibrio parahaemolyticus represents a substantial economic and public health concern; however, elucidating its virulence mechanisms has been significantly impeded by its inherent resistant to genetic manipulation, primarily attributed to sophisticated immune defense systems including restriction-modification (R-M) modules, CRISPR-Cas systems, standalone DNases, and DdmDE systems. Paradoxically, while genetic modification is essential for overcoming these barriers, the very barriers themselves obstruct DNA introduction. Our investigation focused on the V. parahaemolyticus X1 strain, where initial plasmid transformation attempts proved unsuccessful. However, low-efficiency conjugation allowed knockout of defense genes, thereby silencing the host’s defense mechanisms. Our findings revealed a standalone DNase, Vpn, as the predominant obstacle to foreign DNA entry in the X1 strain, while a DdmDE system executes elimination of invaded plasmids. Leveraging these insights, we created the V. parahaemolyticus X2 strain via sequential depletion of the Vpn nuclease and the DdmDE system. Capitalizing on the bacterium’s exceptional growth rate, characterized by a generation time of approximately 10.5 min, we established a highly efficient molecular cloning platform capable of creating a new plasmid construct within a single day. This work not only presents a strategic framework for genetic manipulation of previously recalcitrant bacterial species but also underscores the potential of fast-growing marine bacteria as promising candidates for next-generation biotechnological applications.

|
Scooped by
mhryu@live.com
January 10, 4:39 PM
|
Single-cell resolution studies have transformed our understanding of microbial systems, revealing substantial cell-to-cell heterogeneity and complex dynamic behaviors. This review describes recent advances in using optogenetics, where light-sensitive proteins control cellular processes, to investigate microbial behavior at the individual cell level. We discuss studies where optogenetic approaches have enabled high-resolution analysis of properties such as relative cell positioning, subcellular localization, morphology, and gene expression dynamics. In addition, we highlight emerging feedback and event-driven control methods that dynamically modulate cellular states using light signals. By leveraging light's unique capabilities for spatial and temporal manipulation, researchers can now probe cellular characteristics with unprecedented precision. We anticipate significant advances as researchers introduce more sophisticated dynamically patterned light signals for single-cell microbial research.

|
Scooped by
mhryu@live.com
January 10, 4:34 PM
|
Magnetotactic bacteria (MTB) utilize magnetosomes to align passively with Earth's magnetic field. Magnetic alignment, coupled with flagellar motility and aerotaxis, enables MTB to perform magneto-aerotaxis-a strategy that constrains their movement to a one-dimensional trajectory along geomagnetic field lines, optimizing their search for low-oxygen niches in aquatic environments. Beyond axially constrained movement, environmental MTB isolates exhibit a hemispherically determined swimming polarity-favoring either magnetic north or south-that has been suggested to facilitate descent into oxygen-depleted zones. However, a direct quantitative evaluation of how matching swimming polarity influences navigation toward low-oxygen environments has remained elusive. Here, we employed microcapillary assays to assess the functional significance of polar magneto-aerotaxis in the model organism Magnetospirillum gryphiswaldense. We found that a magnetic field configuration matching the predominant swimming polarity of the population results in an up to 4-fold increased peak intensity of the aerotactic band compared to populations with non-matching polarity. Competition assays using fluorescently labeled north- and south-seeking populations confirmed that congruence between swimming polarity and magnetic field orientation markedly improves aerotactic band formation in oxygen gradients. Alongside our main findings, we noted biomagnetism-independent phototactic responses integrated with aerotaxis, driving collective unidirectional migration along the oxygen gradient. Our results provide direct evidence that matching swimming polarity with the magnetic field confers a clear advantage in navigating oxygen gradients. These findings reinforce the role of the geomagnetic field in shaping MTB behavior and highlight the adaptive value of magnetotactic swimming polarity in environmental navigation.

|
Scooped by
mhryu@live.com
January 10, 3:58 PM
|
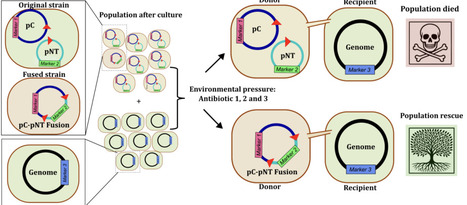
Plasmids are key drivers of bacterial adaptation, yet the mechanisms that generate their diversity remain poorly understood. Here, we show that mobile genetic elements (MGEs) orchestrate a fusion-deletion life cycle that drives the emergence of multireplicon, multidrug-resistant plasmids in Staphylococcus aureus. Large-scale genomic analyses reveal that multireplicon plasmids are widespread and strongly enriched in transposases. Using experimental assays, we demonstrate that rare MGE-mediated fusion events, via homologous recombination or transposition, combine distinct plasmids into single multireplicon elements, expanding gene content and transfer potential. Antibiotic pressure selectively enriches these fused plasmids, rescuing bacterial populations under stress, whereas opposing selective forces, including phage predation, favor deletion derivatives that preserve essential functions and transmissibility. This cyclical process generates dynamic plasmid repertoires with conserved backbones and diverse accessory modules. We propose that MGE-driven fusion-deletion cycles represent a general principle of plasmid evolution, explaining the rapid emergence and persistence of multidrug-resistant plasmids across bacterial pathogens.

|
Scooped by
mhryu@live.com
January 10, 3:41 PM
|
Aptamers are highly selective nucleic acid ligands that overcome many challenges of antibodies, including structural instability and high production costs. The implementation of aptamers into routine research and commercial applications has advanced steadily, though it has been tempered by the limitations of existing discovery methods, which remain relatively slow and low-throughput. SELEX, the gold-standard method for aptamer discovery, has played a pivotal role in the field. As the demand for faster and more tailored solutions grows, there is increasing recognition of the value of newer approaches that can expedite the discovery and optimization of aptamers for specific applications. This review discusses recent advances in the methods used for aptamer engineering, including both wet lab methods performed in vitro, and dry lab methods performed in silico. Applications of engineered aptamers described in recent literature and discuss new developments that could further lead to innovative new applications of aptamers for use both within and outside of the laboratory are discussed. Within the laboratory, these applications include (but are not limited to) target-binding assays and integration into analytical nanomaterials, and outside of the laboratory, diagnostics and biosensing, which will be discussed at length.

|
Scooped by
mhryu@live.com
January 10, 3:19 PM
|
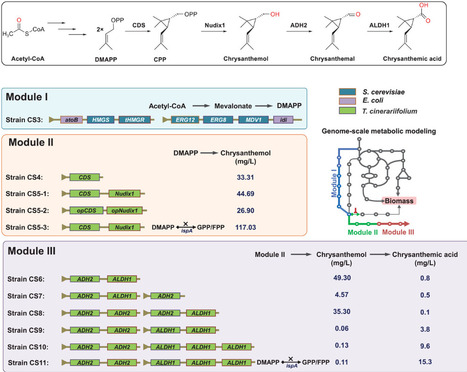
Chrysanthemic acid is an unconventional monoterpene moiety of the natural pesticide pyrethrins with notable anti-insect activity, making its industrial biosynthesis a promising avenue for sustainable agriculture. Here, an E. coli cell factory is designed and build for highly efficient chrysanthemic acid production guided by Genome-scale metabolic models (GEM). The biosynthetic pathway is reconstructed and simulated the metabolic changes caused by exogenous modules are simulated. A key metabolic branch point catalyzed by ispA is identified by this model, and inhibiting its expression using synthetic small RNA redirected the metabolic flux, resulting in the titers of precursor chrysanthemol and chrysanthemic acid increasing by 162% and 59%, respectively. The effect of the expression level of downstream dehydrogenases on chrysanthemic acid titer is also predicated using GEM, and the further optimization of copy number for dehydrogenase genes led to a notably 570% increase in chrysanthemic acid titer experimentally. By integrating the debranching strategy with copy number optimization, a record chrysanthemic acid titer 141.78 mg L−1 is achieved in a bioreactor. The work seamlessly integrated in silico modeling optimization with wet-lab practices that significantly enhance target metabolite titer through metabolic network engineering, offering a new route for constructing efficient cell factories for natural bioproducts.

|
Scooped by
mhryu@live.com
January 10, 3:02 PM
|
Respiratory burst oxidase homolog D (RBOHD)-dependent reactive oxygen species (ROS) in Arabidopsis are well known to suppress pathogen colonization, but their influence on beneficial microbes remains unclear. Here, we found that the beneficial rhizobacterium Pseudomonas anguilliseptica was significantly less enriched in the rhizosphere of rbohD mutants than in that of wild-type plants. Conversely, elevated rhizosphere ROS levels, either triggered by pretreatment with pathogenic Dickeya solani bacteria or caused by mutations in ROS scavenging genes (e.g., in apx1 and cat2 mutants), promoted the rhizosphere recruitment of P. anguilliseptica. This promoting effect was abolished by catalase treatment. In situ microfluidic chemotaxis assays further revealed that P. anguilliseptica exhibits a chemotactic response to low concentrations of hydrogen peroxide ( ≤ 500 nM), accompanied by upregulated expression of chemotaxis- and motility-related genes. Notably, inoculation of P. anguilliseptica effectively suppressed D. solani-induced disease symptoms, and this protective effect was attenuated by catalase treatment. Collectively, these findings reveal a previously unrecognized role of ROS in recruitment beneficial microbiota to enhance plant growth and suppress disease symptoms.
|

|
Scooped by
mhryu@live.com
Today, 1:12 AM
|
Improving the photosynthetic enzyme Rubisco is a key target for enhancing C3 crop productivity, but progress has been hampered by the difficulty of evaluating engineered variants in planta without interference from the native enzyme. Here, we report the creation of a Rubisco-null plant Nicotiana tabacum platform by using CRISPR-Cas9 to knock out all 11 nuclear-encoded small subunit (rbcS) genes. Knockout was achieved in a line expressing cyanobacterial Rubisco from the plastid genome, allowing the recovery of viable plants. We then developed a chloroplast expression system for coexpressing both large and small subunits from the plastid genome. We expressed two resurrected ancestral Rubiscos from the Solanaceae family. The resulting transgenic plants were phenotypically normal and accumulated Rubisco to wild-type levels. Importantly, kinetic analyses of the purified ancestral enzymes revealed they possessed a 16 to 20% higher catalytic efficiency (kcat,air/Kc,air) under ambient conditions, driven by a significantly faster turnover rate (kcat,air). We have demonstrated that our system allows robust in vivo assessment of novel Rubiscos and that ancestral reconstruction is a powerful strategy for identifying superior enzymes to improve photosynthesis in C3 crops.

|
Scooped by
mhryu@live.com
January 11, 11:19 PM
|
Soil biodiversity is a key driver of ecosystem function, including nutrient cycling, organic-matter decomposition, plant productivity, climate regulation and pathogen control (with subsequent effects on animal, human and plant health). A foundational review in 2014 described the functional role of soil biodiversity in ecosystems, but our understanding of the relationship between soil biodiversity and ecosystem functioning has deepened over the past decade. In this Review, we highlight progress in the field, discuss the approaches and methodological advances that have enabled this progress, and identify emerging research questions. Although the spatiotemporal patterns and community dynamics of soil communities are becoming well understood, topics with important knowledge gaps include the climate feedback effects of soils, the ecology of urban soils and the development of soil health indicators. Global collaborative networks, linking existing databases, and monitoring soil biodiversity and ecosystem functioning are important ways to address these knowledge gaps. By considering the relationships between soil biodiversity and ecosystem functioning we can connect small-scale interactions among plants, microorganisms and animals to ecosystem services and planetary sustainability. Soil biodiversity is poorly studied compared to aboveground biodiversity, but is an important driver of ecosystem functioning. This Review discusses advances in research into the relationships between soil biodiversity and ecosystem functioning, highlighting that integrative and causal study approaches will be needed to fill the gaps in our understanding.

|
Scooped by
mhryu@live.com
January 11, 12:15 PM
|
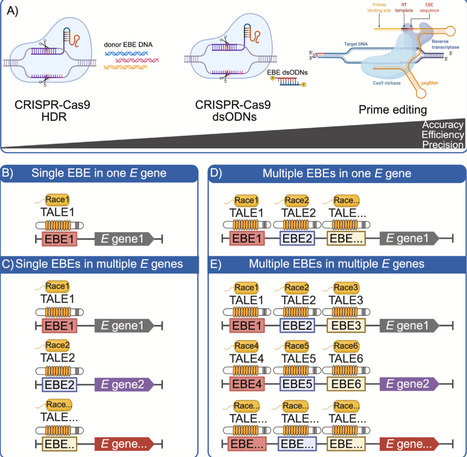
Bacterial type III effector proteins, particularly transcription activator-like effectors (TALEs) secreted by Xanthomonas spp., play critical roles in pathogen–host dynamics. While TALEs facilitate bacterial infections, they also possess vulnerabilities that plants and scientists can exploit to develop mechanisms of resistance. This review encompasses the characteristics and functions of TALEs, examining both their virulence and avirulence roles, and the host plants’ counter-strategies. We highlight advancements in genome editing technologies aimed at combating TALE-dependent plant diseases, with a focus on bacterial blight and leaf streak of rice, but also including bacterial blights of cotton and cassava, and citrus canker. Additionally, we share perspectives on various strategies and approaches for applying genome editing tools to improve disease resistance traits in crop breeding.

|
Scooped by
mhryu@live.com
January 11, 12:02 PM
|
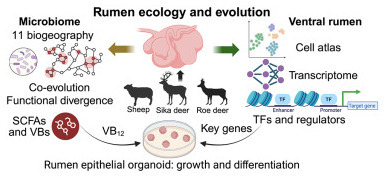
Ruminants thrive in diverse ecosystems by leveraging their rumen microbiome to ferment fibrous plants. However, the spatial biogeography of rumen microbiome and the genetic diversity of the ventral rumen epithelium remain unknown. Here, we present a multi-omics study in roe deer, sika deer, and sheep, integrating region-resolved microbiome and metabolome across 11 ruminal sacs, as well as single-cell RNA sequencing (scRNA-seq), assay for transposase-accessible chromatin using sequencing (ATAC-seq), and bulk RNA sequencing (RNA-seq) of ventral epithelium. We reveal species-specific microbial compositions and metabolic capacities contributing to differences in short-chain fatty acid and vitamin B production. We uncover functional divergence, genomic specialization, and metabolic changes across the microbiome of distinct ruminal sacs. Single-cell profiling reveals changes of immune responses and structural remodeling of the ruminal ventral epithelium. We demonstrate that vitamin B12 promotes epithelial growth and we identify genes enhancing stem cell differentiation. Our results highlight variation in microbial ecology and epithelial architecture among three ruminant species, offering insights to improve livestock productivity.

|
Scooped by
mhryu@live.com
January 11, 11:49 AM
|
Dairy industry generates significant effluent streams, comprising wastewaters and nutrient-rich by-products such as cheese whey, which pose environmental challenges for disposal, but also offer valorisation opportunities. Microalgae cultivation on whey represents a sustainable biotechnological approach that couples biomass production with effluent treatment. This study aimed at optimizing microalgal cultivation using ricotta cheese whey as substrate. Eighteen microalgal strains, mainly belonging to Scenedesmus and Chlorella, were initially screened for their ability to grow on whey. Selected strains were further tested in 300-mL photobioreactors under different management options: Low Whey Load (LWL), High Whey Load (HWL), Salinity and Micronutrient (S&M), and Moderate Whey Load (MWL) trials. Results showed that most of the tested strains grew better when cultivated on diluted whey than on standard synthetic media, with increases from 56 to 590% during the first days, and with high removal efficiencies. Several issues were identified during the trials, including nutrient shortage, salinity increase, micronutrients deficiency and potential inhibitory effects of whey compounds, which have hindered long term cultivation of microalgae on whey. In MWL trial, optimization of whey concentration (12% v/v), replenishment strategy (30% of the culture volume every 3-days), and micronutrient supplementation overcame these issues extending the cultivation period and improving biomass yield (203.6 billion cells L-1 whey-1 with the best strain). Overall, this work underscores the importance of carefully optimize microalgal cultivation management to maximise exploitation of dairy by-products and minimize growth-limiting effects of whey accumulation, thus offering a promising starting point for large-scale applications of whey in microalgae cultivation.

|
Scooped by
mhryu@live.com
January 11, 11:27 AM
|
Recent advances in high-throughput sequencing, imaging, and phenotyping have carried plant science into the era of ‘big data.’ Complex, multi-scale datasets provide new opportunities to uncover plant molecular mechanisms with a level of detail previously unachievable. Fully exploiting this complexity requires integrating advanced statistics, computational modeling, and artificial intelligence (AI). This mini-review offers guidance on how the combination of AI and mechanistic models is transforming temporal, image-based, and spatial omics data into detailed predictions of robust plant traits. In addition, embedding physical principles into AI models can enhance interpretability and strengthen their biological grounding, leading to more realistic representations of plant inner workings. Together, these advances are reshaping plant science by turning ‘big data’ into deep insights, thus greatly enriching our understanding of plant growth, adaptation, and environmental responses.

|
Scooped by
mhryu@live.com
January 10, 4:43 PM
|
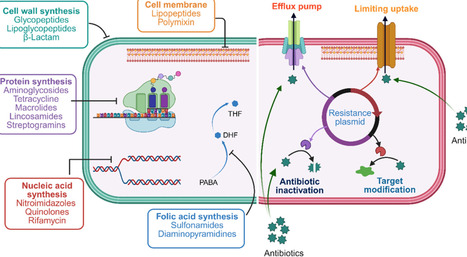
Antibiotics have revolutionized human health by significantly reducing morbidity and mortality associated with bacterial infections. Antibiotics exert bactericidal or bacteriostatic effects through inhibition of cell wall synthesis and disruption of cell membrane integrity, inhibition of protein, nucleic acid synthesis, and other metabolic pathways. Despite their remarkable success since the mid-20th century, antimicrobial resistance (AMR) has emerged as a major global health concern, undermining current treatments and complicating infection management. Key drivers of AMR include the overuse and misuse of antibiotics in clinical settings as well as bacterial adaptations such as genetic mutations and horizontal gene transfer. Mechanistically, these changes can lead to enzymatic inactivation of antibiotics, modification of drug targets, changes in permeability, and active efflux of antimicrobial agents. As resistance rises, antibiotic discovery and development have lagged, creating an urgent need for novel therapeutic strategies and chemical scaffolds. This review examines the antibiotic mechanisms and antibiotic evasion strategies, highlighting genetic and omics approaches used to identify high-priority targets for future drug discovery.

|
Scooped by
mhryu@live.com
January 10, 4:37 PM
|
Bacteria survive hostile conditions in clinically relevant conditions by shutting down protein synthesis, but how they restart growth remains poorly understood. Here, we use an E. coli delta-rimM strain, which exhibits a prolonged growth arrest, as a model to investigate how bacteria recover from this arrested state and restore protein synthesis. RimM is a conserved ribosome maturation factor for the 3'-major (head) domain of the 16S rRNA within the bacterial 30S subunit. The loss of RimM causes a significantly longer delay in recovery than other 30S maturation factors, including RbfA - the presumed primary factor in 30S maturation. Cryo-EM analysis of delta-rimM ribosomes revealed a delayed recruitment of ribosomal proteins to the 30S head domain and increased occupancy of the initiation factors IF1 and IF3, as well as recruitment of the silencing factor RsfS to the 50S subunit. These coordinated changes provide a safeguarding mechanism to block the assembly of premature 70S ribosomes. Notably, while the delayed 30S assembly in delta-rimM reduces the activity of global protein synthesis during the recovery phase, bacteria attempt to compensate for this deficiency by producing higher levels of the ribosomal machinery, indicating a programmatic change in energy allocation to generate the ribosome machinery. These findings highlight the importance of the RimM-assisted assembly of the ribosomal head domain for bacterial recovery from growth arrest.

|
Scooped by
mhryu@live.com
January 10, 4:24 PM
|
Microbial communities often rely on metabolic cooperation, especially under nutrient limitation, but how antibiotics influence these interactions remains unclear. Here, we show that exposure to chloramphenicol, a ribosome-targeting bacteriostatic antibiotic, promotes cooperation in auxotrophic Escherichia coli consortia. Using a proteome-partition model, we find that chloramphenicol-induced growth inhibition elevates levels of the alarmone ppGpp, shifting proteome allocation toward amino acid biosynthesis and export. This regulatory response enhances cross-feeding and supports cooperative growth, even under antibiotic stress. Laboratory experiments confirm that, under low amino acid availability, co-cultures of leucine and phenylalanine auxotrophs display greater tolerance to chloramphenicol than monocultures. Both our theoretical and experimental results reveal a feedback loop in which antibiotic stress strengthens metabolic interdependence, buffering growth inhibition and enhancing community-level tolerance.

|
Scooped by
mhryu@live.com
January 10, 3:57 PM
|
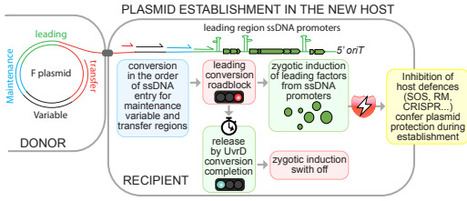
The rapid global spread of antibiotic resistance is mediated by conjugative plasmids, yet how these elements establish immediately after entering new bacterial hosts remains poorly understood. Here we show that plasmids actively coordinate DNA processing with early gene expression to promote their own establishment. Using single-cell measurements of plasmid conversion dynamics, we demonstrate that the F plasmid selectively delays complementary strand synthesis within its leading region, creating a transient single-stranded DNA window that prolongs zygotic induction. This delay is imposed by a single-stranded DNA promoter that forms a structural roadblock to complementary strand DNA synthesis and is resolved by the host helicase UvrD. By extending the single-stranded state of the leading region, plasmids enhance early expression of protective genes that counter host defence responses and promote survival during entry, particularly in non-isogenic hosts. Together, these findings identify early post-entry regulation as a critical determinant of plasmid establishment and reveal how coupling DNA conversion to transient protective gene expression enables conjugative plasmids to overcome host barriers and disseminate antibiotic resistance.

|
Scooped by
mhryu@live.com
January 10, 3:26 PM
|
In recent years, DNA origami technology has advanced rapidly as a groundbreaking method for nanomanufacturing. This technology takes advantage of the unique base-pairing characteristics of DNA, and has significant advantages in constructing spatially ordered and programmable nanostructures. This capability aligns with synthetic biology's core principle of mimicking, extending, and reconstructing natural biological processes by modularly assembling artificial systems. This article provides a comprehensive overview of DNA origami's innovative applications across various domains, including cell membrane surfaces, intercellular communication, intelligent biosensing, and precise gene editing, progressing from the extracellular to the intracellular environment. Finally, this review highlights the synergistic interaction between this technology and cell-free synthetic biology, achieved through the integration of in vitro assembly and cellular regulation, thereby opening new pathways for the rational design of artificial life systems.

|
Scooped by
mhryu@live.com
January 10, 3:15 PM
|
Inspired by the natural ability of bacteriophages to deliver genetic material directly into host cells, we employed a bottom-up approach to construct a multifunctional synthetic DNA origami needle-like structure. This origami is functionalized with trastuzumab antibodies, cholesterol, protective polymers, and two dyes, which together enable selective targeting and insertion into SKBR3 cancer cells. A disulfide-linked dye payload was attached to the apex of the needle, allowing controlled release in the cytoplasm triggered by the high intracellular glutathione concentration. Real-time tracking of the payload confirmed both successful targeting of the origami structure and subsequent direct cytosolic delivery. By mimicking fundamental mechanisms of bacteriophages, we propose that this artificial needle structure can serve as a prototypical device for the targeted delivery of small-molecule drugs directly into the cytosol.
|
 Your new post is loading...
Your new post is loading...



























now bft https://doi.org/10.1093/bib/bbae409