 Your new post is loading...

|
Scooped by
mhryu@live.com
Today, 1:32 AM
|
Worsening global warming, the demand for sustainable chemicals and foods, and the societal imperative to enable healthier and longer lives are redefining the mission of bioengineering. Over the next decade, systems metabolic engineering—where systems-level understanding guides metabolic design and is empowered by synthetic biology, rapid laboratory evolution, and AI—will become an essential approach for addressing these intertwined global challenges. A central challenge is the decarbonization of chemical and materials manufacturing. Systems metabolic engineering will enable microorganisms to transform renewable biomass, industrial wastes, and even CO₂ into bulk and fine chemicals, polymers, and fuels with performance metrics that rival or surpass those of petrochemical processes. The convergence of genome-scale metabolic models, systematic identification of engineering targets and regulatory strategies, and AI-assisted pathway and enzyme design will allow cellular metabolism to be rationally designed, rewired, and optimized to maximize carbon efficiency, redox balance, and energy utilization under industrially relevant conditions. Beyond chemicals and materials, systems metabolic engineering will reshape the production of food and health molecules. Engineered microbes will provide sustainable routes to alternative proteins, functional lipids, vitamins, and structurally complex natural products that are otherwise constrained by land use, climate vulnerability, or low natural abundance. Ultimately, systems metabolic engineering will evolve from strain construction toward AI- and data-driven autonomous biomanufacturing platforms that learn, adapt, and scale, forming foundations for a low-carbon bioeconomy.

|
Scooped by
mhryu@live.com
Today, 1:22 AM
|
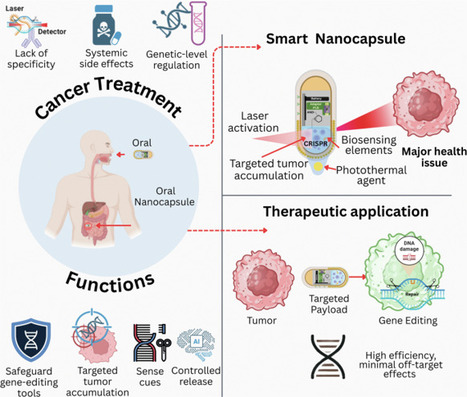
Current therapeutic techniques for cancer often lack specificity. They also cause systemic toxicity and lack genetic control. Thus, cancer ranks among the most complex and crucial global health issues. The novel concept of smart nano-capsules is discussed in this Perspective. These oral medications modify genes using CRISPR technology and integrate biosensing and laser-guided activation to enable more personalized cancer therapies. The creation of these versatile nanocapsules is driven by three objectives. First, they aim to enable controlled gene editing in the gastrointestinal tract. Second, they deliver treatments to specific target areas. Third, they detect tumors in real time. Nanocapsules equipped with biosensing components provide microenvironmental input. An external laser can trigger the release of light-absorbing agents. Moreover, these features reduce off-target effects and allow spatiotemporal precision, the enteric-coated architecture ensures oral stability. Surface functionalization enhances selective tumor accumulation. AI-guided control algorithms can manage diagnostic interpretation and activation. The CRISPR-based cancer medicines offer the potential for improved safety, specificity, and translational use in the future. Combining advanced nanotechnology, gene editing, and AI-guided control could create innovative solutions.

|
Scooped by
mhryu@live.com
Today, 1:17 AM
|
Bacteroides thetaiotaomicron is a model Bacteroidota of the healthy human gut microbiota and a specialist in glycan utilization. Like other Bacteroides, B. theta has many highly regulated polysaccharide utilisation loci (PUL) that encode outer membrane (OM) TonB-dependent transporters (SusC), closely associated ''lid'' lipoproteins (SusD), and additional surface-exposed lipoproteins (SLPs) that bind and partially degrade specific glycans derived from host cells, diet, or other microbiota members. The canonical starch PUL products are thought to form a dynamic complex in the presence of starch. However, other PULs form stable complexes in the absence of substrate (recently named ''utilisomes''), with additional surface lipoproteins tightly associated with the core SusCD complex. In this study, we characterised the B. theta dextran utilisome, with a SusCDdex core and an associated glycoside hydrolase (GHdex) and surface glycan binding protein (SBGPdex). Via X-ray crystallography we solved high-resolution structures of SBGPdex in isolation and SusDdex and GHdex bound to dextran oligosaccharides. We used isothermal titration calorimetry (ITC) to quantify ligand binding of wild type and mutant SLPs. We further used single particle cryo-EM of the catalytically inactive dextran utilisome to visualize open and closed states of the complex. Three occupied dextran binding sites were observed across SusCdex, SusDdex and GHdex, with substrate observed in both open and closed states of SusDdex. 3D variability analysis showed a minority of particles in the process of SusDdex lid closure. Together our work defines commonalities and differences across utilisomes dedicated to the import of simple glycans.

|
Scooped by
mhryu@live.com
Today, 1:01 AM
|
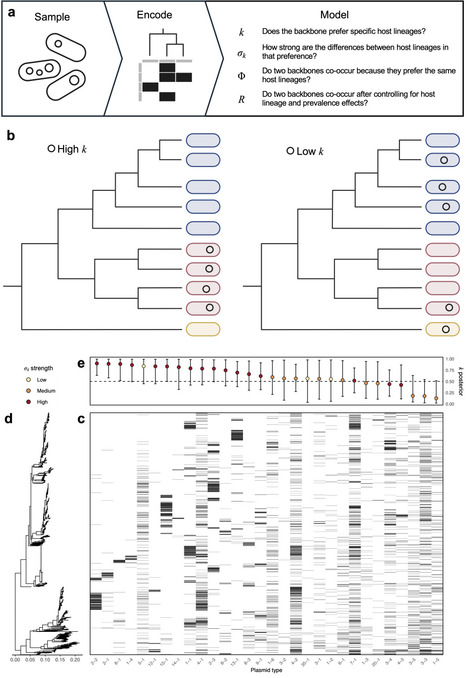
Conjugation mediates plasmid transfer between bacterial species, driving the horizontal spread of traits like antibiotic resistance. However, genomic and experimental evidence indicates that many conjugative plasmids are restricted to particular host lineages despite broad theoretical host ranges. Using 4,281 plasmids from an epidemiologically coherent sample of 1,739 E. coli bloodstream infection isolates, we quantify the host-lineage associations of 30 plasmid backbones and assess how these associations structure plasmid co-occurrence. To achieve this, we develop a novel Bayesian modelling framework that separates genuine backbone-backbone associations from patterns arising due to shared host ancestry or abundance. First, we find that conjugative backbones exhibit stronger host-lineage restriction than mobilisable backbones, and that restriction increases with plasmid size independently of mobility class. Comparison with global plasmid diversity shows that the most host-lineage restricted backbones remain restricted beyond the studied population, whereas the least restricted backbones span a mean of seven host species. Next, after accounting for host phylogeny and abundance, we find that two thirds of backbone pairs show no strong association or avoidance; however, backbones sharing host lineages co-occur more frequently than expected. Lastly, we identify a clique of strongly associated mobilisable backbones that appear to exploit a shared set of lineage-restricted conjugative partners. A mathematical model demonstrates that host-lineage restriction of conjugative backbones, together with specificity in conjugative-mobilisable transfer, is sufficient to generate clustering among mobilisable plasmids. Collectively, our findings reframe conjugation as a mechanism that promotes within-lineage persistence and shapes plasmid community structure, with potentially important consequences for the accumulation of resistance and virulence determinants.

|
Scooped by
mhryu@live.com
Today, 12:27 AM
|
Self-organizing spatial patterns are ubiquitous in microbial ecosystems, yet their sensitivity to environmental conditions remains poorly understood. Understanding spatial pattern sensitivity is particularly relevant for surface-associated microbial systems, as their functioning depends on how different cell-types self-organize across space as a consequence of their traits and environmental conditions. Here, we integrate principles from microbial systems ecology with self-organization theory to understand how environmental conditions and biotic interactions shape the sensitivity of emergent spatial intermixing, which is a critical feature of spatial patterns. Using denitrifying strains of the bacterium Pseudomonas stutzeri that engage in negative (competitive) or positive (cross-feeding) interactions, we demonstrate that spatial intermixing emerging from positive interactions is more sensitive to environmental conditions than that emerging from negative interactions. We further develop and quantify the spatial intermixing strength as a key descriptor of spatial pattern sensitivity, revealing that high short-range dispersal and strong biotic interdependence promote persistent spatial intermixing. Our findings highlight that ecosystem sensitivity to environmental conditions can be inferred from features of emergent spatial patterns, providing a quantitative framework for understanding how environmental and biological factors jointly govern ecosystem assembly and dynamics.

|
Scooped by
mhryu@live.com
February 19, 11:53 PM
|
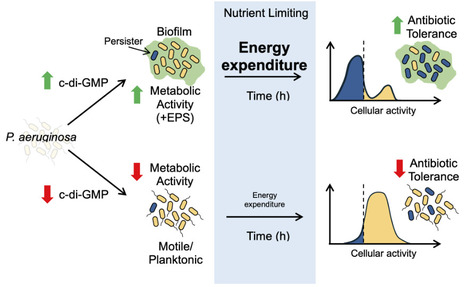
Persisters are a subpopulation of bacterial cells that survive a lethal dose of antibiotic. The failure to treat infections has been linked to the presence of these drug-tolerant cells. The recalcitrant, and often incurable, infection of cystic fibrosis airways by Pseudomonas aeruginosa is attributed to persisters found within aggregate biofilms that contain stationary cells. In all bacteria studied, the fraction of persisters is the highest in stationary populations. However, the level of persisters is unusually low in stationary phase P. aeruginosa, which is unexpected, given the recalcitrance to antibiotic therapy. Here, we set out to investigate the mechanism of P. aeruginosa antibiotic susceptibility in stationary phase. We find that P. aeruginosa is highly active in stationary phase, based on its rate of translation, and this correlates with increased killing compared to E. coli. Growth within alginate beads improves P. aeruginosa stationary cell tolerance to antibiotic treatment, likely due to a subpopulation of low-translating cells. Secondary messenger c-di-GMP regulates biofilm formation, and we find that a population of low-translating persisters also appeared in planktonically growing cells when c-di-GMP production is induced. This phenotype only occurred when exopolysaccharide production was intact, and we propose that the energetic demand of EPS overproduction induces dormancy. The level of energy expenditure appears to determine persistence in P. aeruginosa. In vivo-like growth conditions and a shift to a biofilm lifestyle, mediated by high c-di-GMP production, increase antibiotic tolerance by generating low-energy, dormant persisters.

|
Scooped by
mhryu@live.com
February 19, 11:50 PM
|
Bacillus subtilis is a widely used microbial host for both fundamental research and the industrial production of enzymes and biopharmaceuticals. However, its four known signal peptide–dependent secretion pathways impose inherent limitations on the efficient extracellular expression of heterologous proteins. Here, we identify fructose 1,6-bisphosphate aldolase (FbaA) as a nonclassically secreted protein that achieves high-level expression and efficient extracellular export in B. subtilis. Structural and biochemical data indicate tetrameric FbaA as the predominant secreted form. Site-directed mutagenesis shows that the hydrophobic residue methionine 66 (M66) is critical for tetramer formation; substituting M66 disrupts oligomerization, abolishes secretion, and eliminates FbaA’s ability to mediate the export of heterologous proteins. Together, these results establish that FbaA oligomerization is essential for its nonclassical secretion and that FbaA functions as a modular export element capable of facilitating the secretion of fused heterologous proteins. This work provides mechanistic insight into oligomerization-dependent protein export and offers a promising strategy for engineering efficient secretion systems in B. subtilis.

|
Scooped by
mhryu@live.com
February 19, 11:43 PM
|
Auxotrophs are prevalent in microbial communities, enhancing their diversity and stability—a counterintuitive effect considering their dependence on essential resources from other species. To address the ecological roles of auxotrophs, our study introduced a consumer-resource model (CRM) to capture the complex higher-order interactions within these communities. We also developed an intuitive graphical and algebraic framework, which assesses the feasibility of auxotroph communities and their stability under resource fluctuations and biological invasions. Validated against experimental data from synthetic E. coli auxotroph communities, the model accurately predicted outcomes of community assembly. Our findings highlight the critical role of higher-order interactions and resource dependencies in maintaining the diversity and stability of microbial ecosystems dominated by auxotrophs.

|
Scooped by
mhryu@live.com
February 19, 11:23 PM
|
Riboswitches offer a promising avenue as targets for the development of novel bacterial antibiotics. For this approach to work, it requires that target riboswitches control the expression of essential genes for bacterial survival. However, the small number of essential riboswitch-regulated genes restrains the output of viable novel antimicrobial agents. Here, in an effort to increase the number of riboswitch potential targets, we have assessed the essentiality of Pseudomonas aeruginosa riboswitch-controlled genes in nutrient-limited growth conditions. Using a P. aeruginosa library containing inactivating transposons in riboswitch-controlled genes, we find that the majority of these genes are not required for growth in any tested conditions. However, we find that the inactivation of the S-adenosyl-homocysteine (SAH) hydrolase gene results in the complete lack of P. aeruginosa growth in nutrient-limited conditions. Importantly, bacterial growth is rescued when supplementing S-adenosyl-methionine, which is the precursor of SAH in catalyzed methylation processes. RT-qPCR assays show that although the SAH riboswitch is predicted to control translation initiation, mRNA levels are increased in the presence of SAH, suggesting an additional level of control. Lastly, in vitro structural analyses indicate that the riboswitch undergoes significant rearrangement upon SAH binding and exhibits an affinity toward SAH in the micromolar range. Together, our results provide an approach to discover novel riboswitch targets in nutrient-limited growth conditions and suggest that the SAH-sensing riboswitch constitutes a potential target to inhibit the growth of P. aeruginosa.

|
Scooped by
mhryu@live.com
February 19, 5:39 PM
|
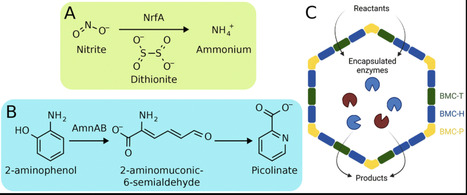
Bacterial microcompartments (BMCs) are protein-based organelles found in diverse bacteria that encapsulate a sequence of enzymes to accelerate specific metabolic pathways and limit toxicity by confining intermediate products. To facilitate catalysis in BMCs, reactants and products must transit across the protein shells, which are made up of multimeric protein tiles. Quantifying the permeability for transport of small molecules across the shell is a key engineering consideration to design novel catalytic microcompartments. We examine the permeability of reactants and products for two reaction pathways, the degradation of 2-aminophenol by AmnAB and the reduction of nitrite to ammonium by NrfA, through a computational lens to quantify permeability for these metabolites at the nanoscale. From Hamiltonian replica exchange umbrella sampling simulations, we determine that the energetic barriers to permeation for the relevant metabolites along both pathways are small, with permeability coefficients in the range of 0.1–4 cm s−1. The high permeabilities calculated for each metabolite through the inhomogeneous solubility diffusion model are corroborated by enzyme activity measurements in vitro that point to enzymatic activity within the shell system. We expand on these findings by quantifying the scale of transport and catalysis that can be supported by in vitro BMC systems. The results highlight the importance of substrate permeability across BMCs within a biological context, and represent steps toward generating synthetic shells or bioengineering novel nanoreactors.

|
Scooped by
mhryu@live.com
February 19, 5:29 PM
|
Cancer immunotherapy is increasingly moving toward personalized, precision-based strategies, with cancer vaccines emerging as a promising approach to reshape treatment. However, despite their potential, current tumor vaccines often yield limited clinical responses and subpar immunogenicity, underscoring the urgent need for innovative delivery systems to enhance immune activation. Bacterial outer membrane vesicles (OMV), which possess natural immunomodulatory properties and impressive engineering flexibility, have attracted attention as versatile platforms for vaccine development and bioengineering applications. This review thoroughly summarizes recent advances in using OMVs to enhance the effectiveness of cancer vaccines. First, we explain the key biological features of OMVs that support their immunotherapeutic potential. Next, we carefully analyze the primary mechanisms by which OMVs enhance immune responses, as well as cutting-edge engineering strategies to improve their safety, immunogenicity, and specificity. Additionally, we discuss the significant challenges that hinder the clinical use of OMV-based cancer vaccines and provide a comprehensive review of current progress and future outlooks. Looking forward, combining artificial intelligence, tumor microenvironment profiling, and neoantigen discovery is expected to drive the development of next-generation, personalized OMV-based immunotherapies. Overall, OMVs stand out as a transformative platform capable of overcoming major obstacles in cancer vaccine development and pushing forward future cancer immunotherapy.

|
Scooped by
mhryu@live.com
February 19, 5:16 PM
|
Phage therapy is an attractive countermeasure to multidrug-resistant pathogens, but clinical deployment is limited by the narrow and poorly predictable host range of most phages. Public repositories now house tens of thousands of phage genomes, yet there is no systematic route for turning this sequence space into designer phages with defined receptor specificities. Here we present a scalable, data-driven framework that converts receptor-binding protein (RBP) diversity into a modular toolkit for programmable phage engineering. Using Klebsiella pneumoniae as a model, we mined 280 non-redundant Przondovirus RBP sequences and resolved them into >50 discrete clusters. Functional screening of the Prz_RBPs from 41 previously uncharacterized Przondovirus phages expanded the number of experimentally validated capsular locus (KL) targets from 14 to 32 in Przondovirus. Each cluster was primarily associated with a dominant KL type, enabling construction of a genotype-to-phenotype map that accurately predicts receptor tropism. The identification of conserved anchor motifs enabled combinatorial pairing of Prz_RBP1 and Prz_RBP2 as plug-and-play modules for programmable phage tropism. Supplying exogenous Prz_RBP2 variants that assemble with Prz_RBP1 further and predictably expands host range, providing broad and tunable coverage. This framework transforms raw genomic diversity into customizable antibacterial agents and offers a general blueprint for precision phage therapy.

|
Scooped by
mhryu@live.com
February 19, 5:07 PM
|
Preozonation is widely used to enhance the effectiveness of coagulation and filtration in algae-laden water treatment, but cyanobacterial cell rupture and the subsequent release of intracellular organic matter and cyanotoxins can increase treatment burdens and pose health risks. In natural waters, cyanobacteria are often surrounded by symbiotic bacteria, whose influence on ozonation performance and underlying mechanisms remains unclear. Herein, we found that axenic filamentous cyanobacteria (Leptolyngbya sp.) exhibited strong resistance to ozonation (0.3 mg L–1, 20 min), whereas the presence of surface-associated bacteria markedly increased the cell rupture rate from 12 ± 6% to 76 ± 2%. Removal of loosely bound extracellular polymeric substances (LB-EPS) significantly reduced ozonation resistance in axenic cyanobacteria but unexpectedly enhanced that of xenic cultures. By integrating reactive oxygen species identification, extracellular metabolomics, and metabolic reconstruction, we demonstrate that surface-colonizing bacteria degrade the algal LB-EPS envelope, releasing metabolites that facilitate hydroxyl radical formation during ozonation, thereby intensifying cell rupture. Our results highlight surface-associated bacteria as a critical yet overlooked factor shaping cyanobacterial responses to preozonation, underscoring the need to re-evaluate ozone application strategies in bloom-impacted waters to minimize cell rupture and byproduct formation.
|

|
Scooped by
mhryu@live.com
Today, 1:26 AM
|
RNA design aims to find a sequence that can fold into a target secondary structure. It can create artificial RNA molecules for specific functions, with wide applications in medicine. It is computationally challenging due to two levels of combinatorial explosion: the exponentially large design space and the exponentially many competing structures per design. Popular methods such as local search cannot keep up with these combinatorial explosions. We instead employ two techniques from machine learning, continuous optimization and Monte-Carlo sampling. We start from a distribution over all valid sequences, and use gradient descent to improve the expectation of an arbitrary objective function. We define novel coupled-variable distributions to model the correlation between nucleotides. We then use sampling to approximate the objective, estimate the gradient, and select the final candidate. Our work consistently outperforms state-of-the-art methods in key metrics including Boltzmann probability and ensemble defect, especially on long and hard-to-design structures. RNA design aims to find a sequence that folds into a target structure. Here, the authors formulate it as a continuous optimization over a coupled-variable distribution and leverage sampling to optimize arbitrary objectives. On Eterna100, it outperforms state-of-the-art methods across key metrics.

|
Scooped by
mhryu@live.com
Today, 1:20 AM
|
Lignin represents a promising and sustainable feedstock for the production of high-value products. However, its heterogeneity and recalcitrance pose a big challenge for efficient depolymerization and upcycling. This review highlights recent advances in microbial lignin valorization, focusing on three key steps: lignin depolymerization, metabolism of lignin-derived aromatic compounds, and valorization to target products. Recent progress in lignin depolymerization is enabling the discovery and optimization of more efficient and broadly specific ligninolytic enzymes, and highlights the critical role of auxiliary enzymes and quinone redox cycling in supporting ligninolytic activity. New catabolic mechanisms, transport systems, and transcriptional regulation networks for both dimeric and monomeric lignin-derived substrates expand our understanding of biological funneling pathways, and they offer valuable tools for designing more efficient microbial biocatalysts and biosensors. Emerging metabolic engineering and adaptive laboratory evolution strategies for creating robust microbial chassis capable of producing diverse value-added products from lignin-derived feedstocks are discussed.

|
Scooped by
mhryu@live.com
Today, 1:11 AM
|
Protein translation is an error-prone process resulting in a random population of altered protein sequences in every cell. Here, we analyzed thousands of publicly available mass spectrometry datasets to detect amino acid misincorporations and quantify error rates in 14 model organisms. We find that overall error rates and the patterns of codon to amino acid error rates correlate across species. We estimate that on average 1-2% of protein molecules in a cell harbor a misincorporation, whereas this proportion can reach 10% for long proteins. Highly expressed and very long proteins have lower error rates, indicating evolutionary selection on codon usage to reduce the cost of translation errors. While both codon-anticodon mispairing and tRNA mischarging contribute to misincorporations, we estimate that ~70% of misincorporation events are due to mispairing. The more frequent an amino acid in the proteome, the more likely it is misincorporated (r = 0.53), likely because frequent amino acids are abundant in the cell, increasing the rate of mischarging, and have abundant tRNAs, leading to increased mispairing. Overall, we find that amino acid and codon usage explain error rates. The conserved patterns of amino acid misincorporations from bacteria to humans suggest universal mechanisms driving translational fidelity.

|
Scooped by
mhryu@live.com
Today, 12:53 AM
|
Fructans are ubiquitous in terrestrial ecosystems, however, these glycans are underexplored in the marine environment. We have discovered that the Antarctic gammaproteobacterium Pseudoalteromonas distincta is highly adapted to the degradation of fructose-containing substrates. This is enabled by proteins encoded in several genomic regions, including a fructan polysaccharide utilization locus (PUL). In addition to a glycoside hydrolase from family 32 (GH32), the fructan PUL encodes two proteins that have been described as specific for the phylum Bacteroidota and were previously unknown for the class Gammaproteobacteria (phylum Pseudomonadota): a glycan-binding SusD-like protein and a SusC-like TonB-dependent transporter (TBDT), which work as a complex in glycan import in Bacteroidota. Proteome, biochemical, sequence, and structural analyses indicate that the SusD-like protein and SusC-like TBDT of P. distincta mediate the uptake of inulin-type fructans, followed by degradation by a periplasmic exo-active GH32. In contrast, P. distincta likely degrades levan-type fructans via an extracellular endo-acting GH32 that is not encoded in the fructan PUL. Comparative genomics identified further SusD-like proteins and SusC-like TBDTs in Gammaproteobacteria, most of which are co-encoded with GH32s, indicative of fructan PULs, and are frequently associated with the marine habitat. Our study thus suggests that SusC/D-like complexes are not exclusive to the phylum Bacteroidota. It further shows that fructans contribute to the marine glycan pool and are targeted by specialized marine communities.

|
Scooped by
mhryu@live.com
Today, 12:25 AM
|
Viruses are obligate parasites that rely extensively on host cellular machinery to complete their life cycles. Therefore, examining the fundamental nature behind how viruses interact with their hosts can teach us more about the pathogenesis of these microbes while also contributing to our overall understanding of essential cell biological processes. In a 2021 mSphere of Influence article, the use of live-cell imaging to uncover a unique mechanism of viral assembly was discussed. In this Full Circle review, we highlight the high-resolution imaging techniques currently revolutionizing virology research and discuss how they can be utilized to advance our ability to identify and interrogate novel virus-host interactions.

|
Scooped by
mhryu@live.com
February 19, 11:51 PM
|
Reduced-size CRISPR systems have become a possible remedy to the delivery and size constraints of the traditional SpCas9 (~ 1368 Å). Recently described small nucleases, including Cas12f (400–700 Å) or CasX (~ 980 Å), along with designed mini-Cas9 versions, can efficiently be used in vivo to edit cells as well as to perform point-of-care diagnostics because of their lower molecular weight and less complex structures. This review will sum up progress in compact Cas protein engineering, guide RNA optimization, and delivery vector miniaturization, and point to their influence in therapeutic gene editing and portable diagnostic platforms. We additionally cover the contemporary issues of interest, such as off-target activity, delivery barriers and regulatory requirements, and future opportunities provided through AI-assisted protein design and synthetic biology. The miniaturized CRISPR technology is bound to substantially transform the translational arena of gene editing and world diagnostics.

|
Scooped by
mhryu@live.com
February 19, 11:47 PM
|
Colorectal cancer (CRC) is heavily influenced by gut microbiota and metabolites such as branched-chain amino acids (BCAAs), which provides essential growth materials for tumors and activates related cancer-promoting pathways. We engineered two E. coli Nissle 1917 strains (ECN)─ECN-Deg and ECN-Tra─to deplete BCAAs in the gut in previous work. In this work, using an AOM/DSS-induced CRC mouse model under the amino acid diet, we found that both strains significantly ameliorated CRC progression, improved survival, restored gut barrier function, and reduced systemic inflammation. Mechanistically, they lowered plasma BCAA levels, suppressed mTOR activation, and modulated retinol and drug metabolism pathways. Our results demonstrate that engineered probiotics targeting BCAAs catabolism can effectively inhibit colorectal tumorigenesis, offering a novel synthetic biology-based approach for cancer therapy.

|
Scooped by
mhryu@live.com
February 19, 11:34 PM
|
The Clostridia are a phylogenetically diverse group of anaerobic, spore-forming bacteria that include species of medical, veterinary and industrial importance. The last two decades have seen major advances in our understanding of Clostridial biology despite the difficulties of anaerobic microbiology and the challenges associated with limited genetic tools. Effort has largely focused on the human pathogen Clostridioides difficile, but many of the methods developed have also proven useful in other species. Here, we present a collection of new genetic tools, including an array of promoters of varying strength, that we have characterized in C. difficile, the food spoilage bacterium Clostridium sporogenes and industrially important Clostridium saccharoperbutylacetonicum. We also present a set of modular plasmids that allow expression of proteins with a variety of tags, including for protein purification and fluorescence microscopy and a method for genetic barcoding of C. difficile to facilitate competitive index experiments. We make these tools available in the hope that they will prove useful to the community in support of our growing understanding of these important bacteria.

|
Scooped by
mhryu@live.com
February 19, 5:46 PM
|
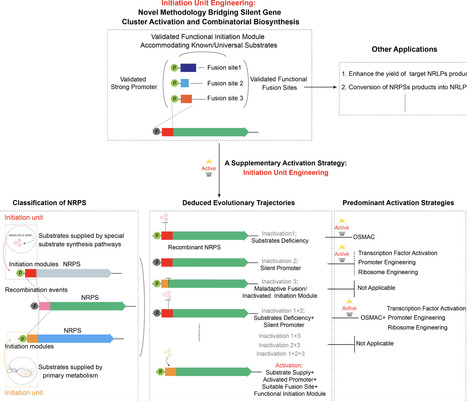
Nonribosomal peptide synthetases (NRPS) represent a valuable yet underexplored resource for producing bioactive natural products. However, most NRPSs remain silenced potentially due to factors such as dysfunction of the initiation unit. The starter condensation (Cs) domain of the initiation unit catalyzes the lipoinitiation of nonribosomal peptides via the incorporation of an N-terminal fatty acyl chain. The concept of initiation unit engineering introduced herein encompasses the replacement of the native initiation unit of NRPSs with a foreign and well-characterized Cs domain-containing initiation unit to activate the NRPS and optimize its expression. This strategy was employed herein to successfully access three of the six previously silent NRPS pathways in Mycetohabitans rhizoxinica HKI 454, a bacterium of the class β-proteobacteria, resulting in the identification of three classes of lipopeptides. This strategy was then extended to access two NRPS pathways in Pseudomonas syringae (γ-proteobacteria) and obtain novel lipopeptides, thereby establishing a feasible complement to existing genome mining strategies for natural product discovery. Furthermore, change of the initiation regions of biosynthetic pathways of nonlipidated chitinimide (β-proteobacteria) and pseudotetraivprolide (γ-proteobacteria) with heterologous Cs-containing initiation units enabled the successful incorporation of fatty acyl chains into the N-terminus of both peptide backbones, launching a workable approach to create artificial lipopeptides. Overall, this study provides a practical strategy for the rational recovery of silent BGCs and introduction of fatty acyl chains into nonribosomal peptides, at least in Proteobacteria, thereby enriching genome mining and combinatorial biosynthesis approaches for accessing the underexplored biosynthetic potential of NRPSs from various bacteria.

|
Scooped by
mhryu@live.com
February 19, 5:33 PM
|
Synthetic microbial communities (SynCom) are microbial consortia with defined taxonomic and functional traits, so that the combination elicits a predictable response under defined conditions. SynComs are artificially designed to enable inter-species metabolic interactions, metabolic division of labor, and ecological interactions that can elicit phenotypes like colonization stability and environmental adaptation. As an applied tool, SynComs have been deployed in diverse contexts, including agriculture, industry, and environmental ecology. This systematic review explores the processes used to construct SynComs, the mechanisms of metabolic interaction between members, and a review of the different ways that SynComs have been applied. We also explore the challenges for SynCom development and application, and future research directions that could overcome these challenges. SynComs are a powerful tool in our arsenal of applied technologies, but research and application are still nascent. While advances have been made, more research is needed to ensure SynCom technologies do not threaten global ecological security. SynCom technology represents a versatile platform for the controlled manipulation of microbial systems, enabling targeted modification of ecological and physiological processes. This emerging field marks a transition from descriptive biology toward a predictive and engineering-driven framework for understanding and shaping living systems.

|
Scooped by
mhryu@live.com
February 19, 5:21 PM
|
Macrophages show great potential for application in cellular immunotherapy, but are limited by immune checkpoints (ICBs). Immune checkpoint inhibitors (ICIs) can effectively block immune escape pathways and alleviate immune suppression in the tumor microenvironment (TME). Here, we constructed hypoxia response bacteria (HRB) that specifically expressed CD47 antibody (aCD47) under hypoxic conditions in the TME, enabling the in situ synthesis of ICIs at the tumor site. In addition, this study further prepared responsive liposomes for encapsulating the STING agonist cGAMP and modified them by covalent attachment to the HRB surface to form the composite material HRB@LC. This composite system can synergistically block CD47-SIRPα-mediated immune escape and activate the STING signal pathway, thereby enhancing systemic antitumor immune responses and significantly improving the efficacy of immunotherapy.

|
Scooped by
mhryu@live.com
February 19, 5:13 PM
|
In Bacillus species, the signal peptide (SP) efficiently guiding the protein secretion is crucial for production, yet there is still a lack of reliable computational tools to accurately predict its efficiency. Therefore, we developed SecEff-Pred, a novel web server that leverages an ESM-2-based predictor. Enhanced by an innovative data simulation strategy and a multitask learning framework, SecEff-Pred accurately predicted the secretion efficiency of signal peptides in Bacillus subtilis. The server demonstrated exceptional performance, achieving prediction accuracies of 85.59% for α-amylase, 81.58% for alkaline xylanase, and 74.68% for cutinase. SecEff-Pred was further validated using phospholipase D (PLD) as a reporter protein, demonstrating high prediction accuracy for signal peptide efficiency (overall 72%), with an accuracy of 80% for “efficient” SPs (corresponding to a maximum PLD activity of 929 U/mL) and 62.50% for “inefficient” SPs. These results confirm the SecEff-Pred is a powerful tool for guiding protein secretion in B. subtilis.
|
 Your new post is loading...
Your new post is loading...



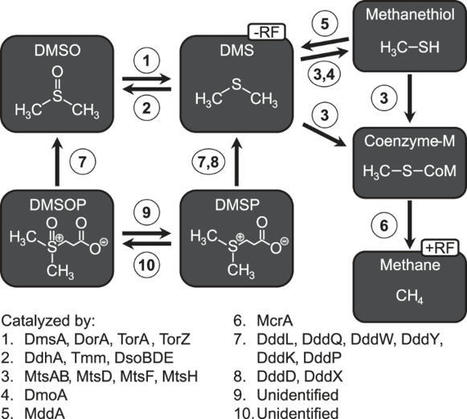
























methane, environmental science