Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
March 31, 2013 1:56 PM
|
The interaction of legumes with N2-fixing bacteria collectively called rhizobia results in root nodule development. The number of nodules formed is tightly restricted through the systemic negative feedback control by the host called autoregulation of nodulation (AON). Here, we report the characterization and gene identification of TOO MUCH LOVE (TML), a root factor that acts during AON in a model legume Lotus japonicus. In our genetic analyses using another root-regulated hypernodulation mutant,plenty, the tml-1 plenty double mutant showed additive effects on the nodule number, whereas the tml-1 har1-7 double mutant did not, suggesting that TML and PLENTY act in different genetic pathways and that TML and HAR1 act in the same genetic pathway. The systemic suppression of nodule formation by CLE-RS1/RS2 overexpression was not observed in the tml mutant background, indicating that TML acts downstream of CLE-RS1/RS2. The tml-1 Snf2 double mutant developed an excessive number of spontaneous nodules, indicating that TML inhibits nodule organogenesis. Together with the determination of the deleted regions in tml-1/-2/-3, the fine mapping of tml-4 and the next-generation sequencing analysis, we identified a nonsense mutation in the Kelch repeat-containing F-box protein. As the gene knockdown of the candidate drastically increased the number of nodules, we concluded that it should be the causative gene. An expression analysis revealed that TMLis a root-specific gene. In addition, the activity of ProTML-GUS was constitutively detected in the root tip and in the nodules/nodule primordia upon rhizobial infection. In conclusion, TML is a root factor acting at the final stage of AON.

|
Scooped by
Jean-Michel Ané
March 27, 2013 12:55 PM
|
The arbuscular mycorrhizal (AM) symbiosis is widespread throughout the plant kingdom and important for plant nutrition and ecosystem functioning. Nonetheless, most terrestrial ecosystems also contain a considerable number of non-mycorrhizal plants. The interaction of such non-host plants with AM fungi (AMF) is still poorly understood. Here, in three complementary experiments, we investigated whether the non-mycorrhizal plant Arabidopsis thaliana, the model organism for plant molecular biology and genetics, interacts with AMF. We grewA. thaliana alone or together with a mycorrhizal host species (either Trifolium pratense or Lolium multiflorum), in the presence or absence of the AMF Rhizophagus irregularis. Plants were grown in a dual-compartment system with a hyphal mesh separating roots of A. thalianafrom roots of the host species, avoiding direct root competition. The host plants in the system ensured the presence of an active AM fungal network. Arbuscular mycorrhizal fungal networks caused growth depressions in A. thaliana of more than 50%, which were not observed in the absence of host plants. Microscopy analyses revealed that R. irregularis supported by a host plant was capable of infecting A. thalianaroot tissues (up to 43% of root length colonized), but no arbuscules were observed. The results reveal high susceptibility of A. thaliana toR. irregularis, suggesting that A. thaliana is a suitable model plant to study non-host/AMF interactions and the biological basis of AM incompatibility.

|
Rescooped by
Jean-Michel Ané
from Plants and Microbes
March 8, 2013 8:48 AM
|
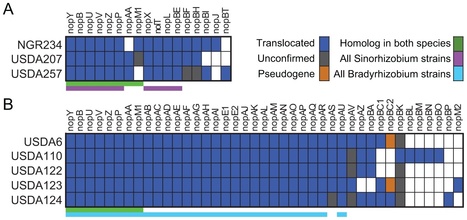
Two diametric paradigms have been proposed to model the molecular co-evolution of microbial mutualists and their eukaryotic hosts. In one, mutualist and host exhibit an antagonistic arms race and each partner evolves rapidly to maximize their own fitness from the interaction at potential expense of the other. In the opposing model, conflicts between mutualist and host are largely resolved and the interaction is characterized by evolutionary stasis. We tested these opposing frameworks in two lineages of mutualistic rhizobia,Sinorhizobium fredii and Bradyrhizobium japonicum. To examine genes demonstrably important for host-interactions we coupled the mining of genome sequences to a comprehensive functional screen for type III effector genes, which are necessary for many Gram-negative pathogens to infect their hosts. We demonstrate that the rhizobial type III effector genes exhibit a surprisingly high degree of conservation in content and sequence that is in contrast to those of a well characterized plant pathogenic species. This type III effector gene conservation is particularly striking in the context of the relatively high genome-wide diversity of rhizobia. The evolution of rhizobial type III effectors is inconsistent with the molecular arms race paradigm. Instead, our results reveal that these loci are relatively static in rhizobial lineages and suggest that fitness conflicts between rhizobia mutualists and their host plants have been largely resolved.
Via Kamoun Lab @ TSL

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
March 6, 2013 8:50 AM
|
Symbiotic nitrogen fixation by rhizobia in legume root nodules injects approximately 40 million tonnes of nitrogen into agricultural systems each year. In exchange for reduced nitrogen from the bacteria, the plant provides rhizobia with reduced carbon and all the essential nutrients required for bacterial metabolism. Symbiotic nitrogen fixation requires exquisite integration of plant and bacterial metabolism. Central to this integration are transporters of both the plant and the rhizobia, which transfer elements and compounds across various plant membranes and the two bacterial membranes. Here we review current knowledge of legume and rhizobial transport and metabolism as they relate to symbiotic nitrogen fixation. Although all legume-rhizobia symbioses have many metabolic features in common, there are also interesting differences between them, which show that evolution has solved metabolic problems in different ways to achieve effective symbiosis in different systems. Michael Udvardi and Philip S. Poole (2013). Annual Review of Plant Biology 64: first posted online March 1.
Via IvanOresnik

|
Scooped by
Jean-Michel Ané
February 28, 2013 8:23 PM
|
Beneficial associations between plants and arbuscular mycorrhizal fungi play a major role in terrestrial environments and in the sustainability of agroecosystems. Proteins, microRNAs, and small molecules have been identified in model angiosperms as required for the establishment of arbuscular mycorrhizal associations and define a symbiotic ‘toolkit’ used for other interactions such as the rhizobia–legume symbiosis. Based on recent studies, we propose an evolutionary framework for this toolkit. Some components appeared recently in angiosperms, whereas others are highly conserved even in land plants unable to form arbuscular mycorrhizal associations. The exciting finding that some components pre-date the appearance of arbuscular mycorrhizal fungi suggests the existence of unknown roles for this toolkit and even the possibility of symbiotic associations in charophyte green algae.

|
Scooped by
Jean-Michel Ané
February 21, 2013 8:53 AM
|
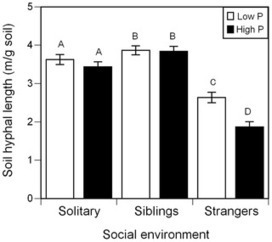
Background: The stability of cooperative interactions among different species can be compromised by cheating. In the plant-mycorrhizal fungi symbiosis, a single mycorrhizal network may interact with many plants, providing the opportunity for individual plants to cheat by obtaining nutrients from the fungi without donating carbon. Here we determine whether kin selection may favour plant investment in the mycorrhizal network, reducing the incentive to cheat when relatives interact with a single network.Methodology/Principal Findings: We show that mycorrhizal network size and root colonization were greater when Ambrosia artemisiifolia L. was grown with siblings compared to strangers. Soil fungal abundance was positively correlated with group leaf nitrogen, and increased root colonization was associated with a reduced number of pathogen-induced root lesions, indicating greater benefit to plants grown with siblings.Conclusions/Significance: Plants can benefit their relatives through investment in mycorrhizal fungi, and kin selection in plants could promote the persistence of the mycorrhizal symbiosis.

|
Rescooped by
Jean-Michel Ané
from Plants and Microbes
February 21, 2013 8:44 AM
|
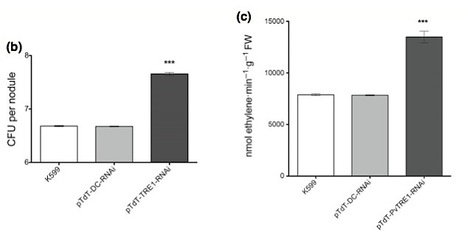
The expression of the promoter of the trehalase gene PvTRE1 fused to GUS is restricted to nodules in Phaseolus vulgaris. Courtesy of Aarón Barraza & Federico Sánchez.
Via Kamoun Lab @ TSL

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
February 20, 2013 8:51 AM
|
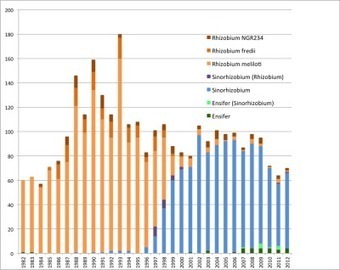
There are many publications on the bacteria that most people know as Sinorhizobium meliloti, S. fredii, and related species. The name of their genus has changed over the years, and I used Web of Science to track the ...
Via IvanOresnik

|
Scooped by
Jean-Michel Ané
February 5, 2013 5:40 PM
|
Legumes control the nitrogen-fixing root nodule symbiosis in response to external and internal stimuli, such as nitrate, and via systemic autoregulation of nodulation (AON). Overexpression of the CLV3/ESR-related (CLE) pre-propeptide-encoding genes GmNIC1 (nitrate-induced and acting locally) and GmRIC1 (Bradyrhizobium-induced and acting systemically) suppresses soybean nodulation dependent on the activity of the nodulation autoregulation receptor kinase (GmNARK). This nodule inhibition response was used to assess the relative importance of key structural components within and around the CLE domain sequences of these genes. Using a site-directed mutagenesis approach, mutants were produced at each amino acid within the CLE domain (RLAPEGPDPHHN) ofGmRIC1. This approach identified the Arg1, Ala3, Pro4, Gly6, Pro7, Asp8, His11, and Asn12 residues as critical to GmRIC1 nodulation suppression activity (NSA). In contrast, none of the mutations in conserved residues outside of the CLE domain showed compromised NSA. Chimeric genes derived from combinations of GmRIC1 and GmNIC1 domains were used to determine the role of each pre-propeptide domain in NSA differences that exist between the two peptides. It was found that the transit peptide and CLE peptide regions of GmRIC1 significantly enhanced activity of GmNIC1. In contrast, the comparable GmNIC1 domains reduced the NSA of GmRIC1. Identification of these critical residues and domains provides a better understanding of how these hormone-like peptides function in plant development and regulation.

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
January 31, 2013 8:56 AM
|
The ability of the photosynthetic Bradyrhizobium strains ORS285 and ORS278 to nodulate soybeans was investigated. While the nod gene-deficient ORS278 strain induced only bumps on soybean roots, the nod gene-containing strain ORS285 formed nitrogen-fixing nodules. However, symbiotic efficiency differed drastically depending on both the soybean genotypes used and the culture conditions tested. Giraud E, Xu L, Chaintreuil C, Gargani D, Gully D, Sadowsky MJ. (2013). Appl Environ Microbiol. Jan 25. [Epub ahead of print]
Via IvanOresnik

|
Rescooped by
Jean-Michel Ané
from Plants and Microbes
January 28, 2013 8:57 AM
|
The goal of this symposium is to bring together researchers working in a wide range of disciplines in plant biology, ecology and evolution in the field of plant interactions with other organisms. The central objective is to discuss and integrate information from different approaches and perspectives in order to create a synthetic framework for understanding these interactions. Our hope is to have a diverse and dynamic group of scientists, who are willing to step out of their disciplinary comfort zone and engage in an effort to participate in discussions spanning the range from molecular approaches to ecosystem implications.
Via Kamoun Lab @ TSL

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
January 24, 2013 8:59 AM
|
Resources from the Sinorhizobium meliloti Rm1021 open reading frame (ORF) plasmid libraries were used in a medium-throughput method to construct a set of 50 overlapping deletion mutants covering all of the Rm1021 pSymA megaplasmid except the replicon region. Each resulting pSymA derivative carried a defined deletion of approximately 25 ORFs. Various phenotypes, including cytochrome crespiration activity, the ability of the mutants to grow on various carbon and nitrogen sources and the symbiotic effectiveness of the mutants with alfalfa were analyzed. This approach allowed us to systematically evaluate the potential impact of regions of Rm1021 pSymA for their free-living and symbiotic phenotypes.
Yurgel SN, Mortimer MW, Rice J, Humann J, Kahn ML. (2013). Appl Environ Microbiol. Jan 18. [Epub ahead of print]
Via IvanOresnik

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
January 12, 2013 12:12 PM
|
Abstract Microevolution and origins of Bradyrhizobium populations associated with soybeans at two field sites (A and B, 280 km apart in Canada) with contrasting histories of inoculation was investigated using probabilistic analyses of six core (housekeeping) gene sequences. These analyses supported division of 220 isolates in five lineages corresponding either to B. japonicum groups 1 and 1a or to one of three novel lineages within the genus Bradyrhizobium. None of the isolates from site A and about 20% from site B (the only site with a recent inoculation history) were attributed to inoculation sources. The data suggest that most isolates were of indigenous origin based on sequence analysis of 148 isolates of soybean-nodulating bacteria from native legumes (Amphicarpaea bracteata and Desmodium canadense). Isolates from D. canadense clustered with B. japonicum group 1, whereas those from A. bracteata were placed in two novel lineages encountered at soybean field sites. One of these novel lineages predominated at soybean sites and exhibited a significant clonal expansion likely reflecting selection by the plant host. Homologous recombination events detected in the 35 sequence types from soybean sites had an effect on genetic diversification that was approximately equal to mutation. Interlineage transfer of core genes was infrequent and mostly attributable to gyrB that had a history of frequent recombination. Symbiotic gene sequences (nodC and nifH) of isolates from soybean sites and native legumes clustered in two lineages corresponding to B. japonicum and B. elkani with the inheritance of these genes appearing predominantly by vertical transmission. The data suggest that soybean-nodulating bacteria associated with native legumes represent a novel source of ecologically adapted bacteria for soybean inoculation. Tang J, Bromfield ES, Rodrigue N, Cloutier S, Tambong JT. (2012). Ecol Evol. Dec;2(12):2943-61. doi: 10.1002/ece3.404. Epub 2012 Oct 22.
Via IvanOresnik
|

|
Scooped by
Jean-Michel Ané
March 31, 2013 1:55 PM
|
Symbiotic nitrogen fixation by intracellular rhizobia within legume root nodules requires the exchange of nutrients between host plant cells and their resident bacteria. While exchanged molecules imply nitrogen compounds, carbohydrates and also various minerals, knowledge of the molecular basis of plant transporters that mediate those metabolite exchanges is still limited. In this study, we have shown that a multidrug and toxic compound extrusion (MATE) protein, LjMATE1, is specifically induced during nodule formation, which nearly paralleled nodule maturation, in a model legume Lotus japonicus. Reporter gene experiments indicated that the expression of LjMATE1 was restricted to the infection zone of nodules. To characterize the transport function of LjMATE1, we conducted a biochemical analysis using a heterologous expression system, Xenopus oocytes, and found that LjMATE1 is a specific transporter for citrate. The physiological role of LjMATE1 was analyzed after generation of L. japonicus RNA interference (RNAi) lines. One RNAi knock-down line revealed limited growth under nitrogen-deficient conditions with inoculation of rhizobia compared with the controls (the wild type and an RNAi line in which LjMATE1 was not suppressed). It was noteworthy that Fe localization was clearly altered in nodule tissues of the knock-down line. These results strongly suggest that LjMATE1 is a nodule-specific transporter that assists the translocation of Fe from the root to nodules by providing citrate.

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
March 17, 2013 7:20 PM
|
Background. Sinorhizobium meliloti is a soil-dwelling alpha-proteobacterium that possesses a large, tripartite genome and engages in a nitrogen fixing symbiosis with its plant hosts. Although much is known about this important model organism, global characterization of genetic regulatory circuits has been hampered by a lack of information about transcription and promoters. Results. Using an RNAseq approach and RNA populations representing 16 different growth and stress conditions, we comprehensively mapped S. meliloti transcription start sites (TSS). Our work identified 17,001 TSS that we grouped into six categories based on the genomic context of their transcripts: mRNA (4,430 TSS assigned to 2,657 protein-coding genes), leaderless mRNAs (171), putative mRNAs (425), internal sense transcripts (7,650), antisense RNA (3,720), and trans-encoded sRNAs (605). We used this TSS information to identify transcription factor binding sites and putative promoter sequences recognized by seven of the 15 known S. meliloti sigma factors sigma70, sigma54, sigmaH1, sigmaH2, sigmaE1, sigmaE2, and sigmaE9). Altogether, we predicted 2,770 new promoter sequences, including 1,302 located upstream of protein coding genes and 722 located upstream of antisense RNA or trans-encoded sRNA genes. To validate promoter predictions for targets of the general stress response sigma factor, RpoE2 (sigmaE2), we identified rpoE2-dependent genes using microarrays and confirmed TSS for a subset of these by 5[prime] RACE mapping. Conclusions. By identifying TSS and promoters on a global scale, our work provides a firm foundation for the continued study of S. meliloti gene expression with relation to gene organization, sigma factors and other transcription factors, and regulatory RNAs. Jan-Philip Schlüter, Jan Reinkensmeier, Melanie J Barnett, Claus Lang, Elizaveta Krol, Robert Giegerich, Sharon R Long and Anke Becker (2013). BMC Genomics 14:156.
Via IvanOresnik

|
Scooped by
Jean-Michel Ané
March 6, 2013 9:04 AM
|
At the conference Forsythe's presentation entitled A Field Study of Specificity in the Arbuscular Mycorrhizal Symbiosis won the prestigious John L. Harley Medal for excellence. The John L. Harley Medal is awarded at each ...

|
Scooped by
Jean-Michel Ané
March 3, 2013 9:44 AM
|
Life demands tradeoffs, and plants are no exception. Virginia wildrye, common on U.S. prairies and rangelands, often plays host to a fungus that helps this grass grow. But the plant pays a price. Researchers have discovered that infected plants produce less pollen than their noninfected counterparts. Instead, the fungus causes the rye grass to make extra seeds, which transmit the fungus to the next generation and new locations. This is the first time a fungus has proven capable of manipulating plant reproduction. The finding highlights the complexity of the relationship between hosts and their guests.

|
Scooped by
Jean-Michel Ané
February 21, 2013 11:27 AM
|

|
Scooped by
Jean-Michel Ané
February 21, 2013 8:45 AM
|
University of Delaware
Janine Sherrier has been named the acting deputy dean for the University of ...

|
Rescooped by
Jean-Michel Ané
from Plants and Microbes
February 21, 2013 8:43 AM
|
Legume–rhizobium interactions have been widely studied and characterized, and the disaccharide trehalose has been commonly detected during this symbiotic interaction. It has been proposed that trehalose content in nodules during this symbiotic interaction might be regulated by trehalase. In the present study, we assessed the role of trehalose accumulation by down-regulating trehalase in the nodules of common bean plants. We performed gene expression analysis for trehalase (PvTRE1) during nodule development. PvTRE1 was knocked down by RNA interference (RNAi) in transgenic nodules of the common bean. PvTRE1 expression in nodulated roots is mainly restricted to nodules. Down-regulation of PvTRE1 led to increased trehalose content (78%) and bacteroid number (almost one order of magnitude). In addition, nodule biomass, nitrogenase activity, and GOGAT transcript accumulation were significantly enhanced too. The trehalose accumulation, triggered by PvTRE1 down-regulation, led to a positive impact on the legume–rhizobium symbiotic interaction. This could contribute to the agronomical enhancement of symbiotic nitrogen fixation.
Via Kamoun Lab @ TSL

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
February 20, 2013 8:36 AM
|
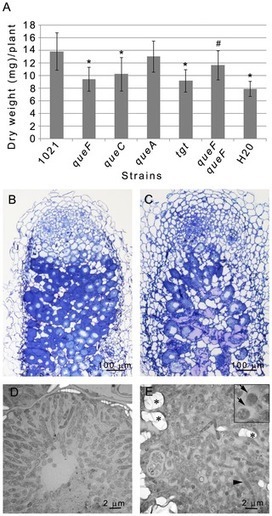
Rhizobia are symbiotic soil bacteria able to intracellularly colonize legume nodule cells and form nitrogen-fixing symbiosomes therein. How the plant cell cytoskeleton reorganizes in response to rhizobium colonization has remained poorly understood especially because of the lack of an in vitro infection assay. Here, we report on the use of the heterologous HeLa cell model to experimentally tackle this question. We observed that the modelrhizobium Sinorhizobium meliloti, and other rhizobia as well, were able to trigger a major reorganization of actin cytoskeleton of cultured HeLa cells in vitro. Cell deformation was associated with an inhibition of the three major small RhoGTPases Cdc42, RhoA and Rac1. Bacterial entry, cytoskeleton rearrangements and modulation of RhoGTPase activity required an intact S. meliloti biosynthetic pathway for queuosine, a hypermodifed nucleoside regulating protein translation through tRNA, and possibly mRNA, modification. We showed that an intact bacterial queuosine biosynthetic pathway was also required for effective nitrogen-fixing symbiosis of S. meliloti with its host plant Medicago truncatula, thus indicating that one or several key symbiotic functions of S. meliloti are under queuosine control. We discuss whether the symbiotic defect of que mutants may originate, at least in part, from an altered capacity to modify plant cell actin cytoskeleton. Marchetti M, Capela D, Poincloux R, Benmeradi N, Auriac MC, Le Ru A, Maridonneau-Parini I, Batut J, Masson-Boivin C. (2013). PLoS One. ;8(2):e56043. Epub 2013 Feb 8.
Via IvanOresnik

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
February 1, 2013 10:45 AM
|
The intensive application of fertilizers during agricultural practices has led to an unprecedented perturbation of the nitrogen cycle, illustrated by the growing accumulation of nitrates in soils and waters, and of nitrogen oxides in the atmosphere. Besides increasing use efficiency of current N fertilizers, priority should be given to put on value the process of biological nitrogen fixation, through more sustainable technologies that reduce the undesired effects of chemical N fertilization of agricultural crops. Wider legume adoption, supported by coordinated legume breeding and inoculation programs are approaches at hand. Also available are biofertilizers based on microbes that help to reduce the needs of N fertilization in important crops like cereals. Engineering in cereals the capacity to fix nitrogen, either by themselves or in symbiosis with nitrogen-fixing microbes, are attractive future options that nevertheless require more intensive and internationally coordinated research efforts. Although nitrogen-fixing plants may be less productive, at some point agriculture must significantly reduce the use of warming (chemically synthesized) N and give priority to the biological nitrogen fixation, if it is to sustain both food production and environmental health for a continuously growing human population. Olivares J, Bedmar EJ, Sanjuan J.(2013). Mol Plant Microbe Interact. Jan 29. [Epub ahead of print]
Via IvanOresnik

|
Rescooped by
Jean-Michel Ané
from Mycorrhizal fungal genomes
January 28, 2013 8:58 AM
|
Arbuscular mycorrhizal fungi (AMF) represent an ecologically relevant and evolutionarily intriguing group of land plant symbionts, which produce multinucleated spores and hyphae that are currently thought to have propagated clonally for over 500 million years. This long-term absence of sex in AMF is a puzzling evolutionary feature that has sparked scientific interest for some time, but a provoking explanation for their successful evolutionary history in the absence of an obvious sexual cycle is that these organisms may have cryptic sex, or a parasexual life cycle, allowing them to recombine alleles and compensate for deleterious mutations. Interestingly, the recent acquisition of large sequence data from many AMF species can finally allow this hypothesis to be tested more extensively. In this perspective, we highlight emerging evidence based on sequence data for the potential of AMF to have sexual reproduction, and propose a number of routes that could be taken to further explore the presence (or absence thereof) of sex in this poorly studied, yet highly relevant, fungal group. Rohan Riley, Nicolas Corradi Fungal Ecology Volume 6, Issue 1, February 2013, Pages 44–49 http://dx.doi.org/10.1016/j.funeco.2012.01.010
Via Stefano Ghignone

|
Rescooped by
Jean-Michel Ané
from Rhizobium Research
January 28, 2013 8:56 AM
|
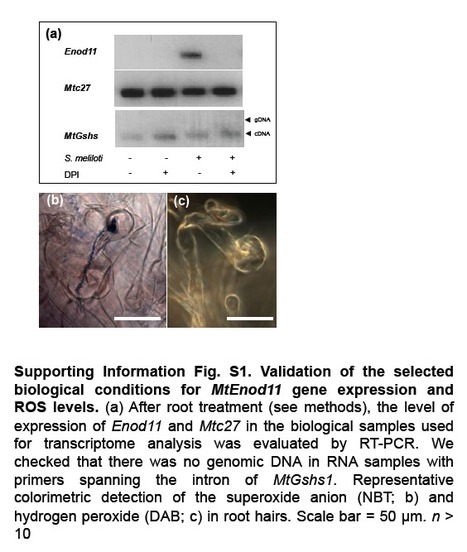
Reactive oxygen species (ROS), particularly hydrogen peroxide (H(2) O(2) ), play an important role in signalling in various cellular processes. The involvement of H(2) O(2) in the Medicago truncatula-Sinorhizobium meliloti symbiotic interaction raises questions about its effect on gene expression. A transcriptome analysis was performed on inoculated roots of M. truncatula in which ROS production was inhibited with diphenylene iodonium (DPI). In total, 301 genes potentially regulated by ROS content were identified 2 d after inoculation. These genes included MtSpk1, which encodes a putative protein kinase and is induced by exogenous H(2) O(2) treatment. MtSpk1 gene expression was also induced by nodulation factor treatment. MtSpk1 transcription was observed in infected root hair cells, nodule primordia and the infection zone of mature nodules. Analysis with a fluorescent protein probe specific for H(2) O(2) showed that MtSpk1 expression and H(2) O(2) were similarly distributed in the nodule infection zone. Finally, the establishment of symbiosis was impaired by MtSpk1 downregulation with an artificial micro-RNA. Several genes regulated by H(2) O(2) during the establishment of rhizobial symbiosis were identified. The involvement of MtSpk1 in the establishment of the symbiosis is proposed. Andrio E, Marino D, Marmeys A, de Segonzac MD, Damiani I, Genre A, Huguet S, Frendo P, Puppo A, Pauly N. (2013). New Phytol. Jan 24. doi: 10.1111/nph.12120. [Epub ahead of print]
Via IvanOresnik

|
Scooped by
Jean-Michel Ané
January 18, 2013 2:39 PM
|
During the early 1980s, Bauer and associates reported that nodulation potential in primary roots of soybean seedlings following inoculation with rhizobia was significantly reduced in developmentally younger regions. They suggested that this phenomenon might be due to a fast-acting regulatory mechanism in the host that prevented excessive nodulation. However, the molecular mechanism of this fast-acting regulatory response remains uncertain. Here, we sought to elucidate components of this regulatory mechanism by investigating the expression of the NSP1and NSP2 genes that encode a GRAS transcription factor required for nodule initiation. First, we confirmed that younger regions of Lotus japonicus roots also show a reduction in nodule numbers in response toMesorhizobium loti. Then, we compared the expression levels of NSP1and NSP2 in developmentally younger regions of primary roots. After inoculation with M. loti, expression of NSP1 was transiently induced whereas that of NSP2 was significantly downregulated 1 day after inoculation. This result implicates that downregulation of NSP2 might cause a fast-acting regulatory mechanism to prevent further nodulation. Next we overexpressed NSP2 in wild type plants. Overexpression resulted in the clustering of nodules in the upper region of the root but strong suppression of nodulation in the lower region. By contrast, overexpression of NSP2 in har1 hypernodulating mutants resulted in an increased number of nodule primordia even in the root tip region. These results indicate that HAR1 negatively regulates NSP2-induced excessive nodule formation in the developmentally younger regions of roots.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...