Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
February 16, 8:03 PM
|
Plants rely on specialized metabolites to defend against herbivores, yet how root-associated symbiotic microbes influence their regulation, coordination, and overall contribution to defense remains widely unknown.
We hypothesized that symbiotic microbes can reprogram specialized metabolic networks, thereby enhancing herbivore resistance, with a particular focus here on polyamine metabolic network. Using the fungal symbiotic endophyte Trichoderma harzianum, tomato, and the herbivore Spodoptera exigua, we combined greenhouse bioassays with molecular, genetic, and metabolomic analyses to investigate how Trichoderma symbiosis reshapes the plant polyamine metabolic network, and affects plant-herbivore interactions.
We found that in the absence of the root symbiosis, herbivory primarily activated polyamine uptake transport and catabolism, reflecting a stress-driven turnover response. Trichoderma symbiosis markedly reconfigured the polyamine network. Symbiosis primed uptake transport and catabolic responses, enhanced polyamine flux through activation of the ornithine decarboxylase pathway, and redirected polyamines into conjugated metabolites with anti-herbivore activity. Genetic analyses confirmed that this metabolic rewiring contributes to Trichoderma-induced resistance to herbivory, linking primary metabolic routes to the accumulation of specialized defense compounds.
Our study highlights root symbionts as key modulators of plant metabolism, showing that specialized metabolite diversity can be shaped by symbiotic interactions.

|
Scooped by
Jean-Michel Ané
February 15, 4:19 PM
|
The seed-to-seedling transition is a key step of plant microbiota assembly, where the seed microbiota meets the soil microbiota. Seed germination triggers profound physiological changes, including exudate release and oxidative bursts, which generate selective pressures shaping microbial survival and interactions. Here, we investigated how host selection and community composition influence bacterial colonization during this transition using a Synthetic Community (SynCom) approach on common bean (Phaseolus vulgaris) seeds. Specifically, we tested the importance of microbial interactions by applying 30 bacterial SynComs spawning a phylogenetic diversity gradient.
Seedling colonization success was primarily determined by strain identity but was strongly modulated by the SynCom context. Strains that were highly effective colonizers when inoculated alone often exhibited reduced transmission within SynComs, indicating that microbial interactions can either inhibit or facilitate colonization. Random forest models confirmed that colonization outcomes could not be predicted from single-strain performance alone. Instead, both phylogenetic relatedness and metabolic similarity among SynCom members emerged as key predictors of strain success, supporting the competition–relatedness hypothesis and highlighting the importance of community-dependent effects in shaping colonization success. Genomic analyses identified microbial traits linked to efficient seedling colonization, notably amino acid transport and metabolism, stress tolerance, ROS detoxification, and biofilm formation, highlighting the strong selective pressures acting during the seed-to-seedling transition. Besides key adaptative traits, our findings underscore the importance of microbial interactions to survive and colonize seedlings, bringing a novel perspective for the successful engineering of seed and seedling microbiota.

|
Scooped by
Jean-Michel Ané
February 12, 8:08 PM
|
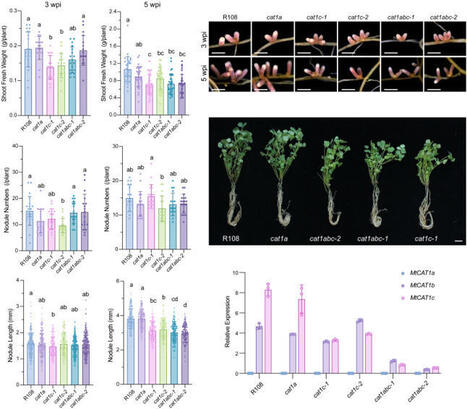
Legumes engage in nitrogen-fixing symbiosis with rhizobia, in which host legumes supply dicarboxylates as a carbon source to rhizobia, while rhizobia reciprocate by providing ammonium to the host plants. Beyond this classical model, accumulating evidence suggests that amino acid exchange is also essential for legume–rhizobium symbiosis. However, it remains unclear whether amino acid transporters are present on the symbiosome membrane (SM) to mediate amino acid exchange during symbiotic nitrogen fixation (SNF). In this study, we identified three amino acid transporters in Medicago truncatula—MtCAT1a, MtCAT1b, and MtCAT1c—which belong to a clade of the plant Cationic Amino acid Transporter (CAT) family known to transport a wide range of amino acids. Notably, MtCAT1b and MtCAT1c are predominantly expressed in infected nodule cells and localize to the SM. Genetic analyses further demonstrate that both MtCAT1b and MtCAT1c are required for amino acid exchange at the SM, with additional evidence indicating that bacteroid metabolism is disturbed in the mutants. Transport assays show that both MtCAT1b and MtCAT1c exhibit broad substrate specificity. Collectively, these findings identify MtCAT1b and MtCAT1c as key mediators of cross-kingdom amino acid exchange, which is essential for maintaining efficient SNF in root nodules.

|
Scooped by
Jean-Michel Ané
February 12, 5:53 PM
|
Symbiotic nutrient exchange between arbuscular mycorrhizal (AM) fungi and their host plants varies widely depending on their physical, chemical, and biological environment. Yet dissecting this context dependency remains challenging because we lack methods for tracking nutrients such as carbon (C) and phosphorus (P). Here, we developed an approach to quantitatively estimate C and P fluxes in the AM symbiosis from comprehensive network morphology quantification, achieved by robotic imaging and machine learning based on roughly 100 million hyphal shape measurements. We found that rates of C transfer from the plant and P transfer from the fungus were, on average, related proportionally to one another. This ratio was nearly invariant across AM fungal strains despite contrasting growth phenotypes but was strongly affected by plant host genotype. Fungal phenotype distributions were bounded by a Pareto front with a shape favoring specialization in an exploration–exploitation trade-off. This means AM fungi can be fast range expanders or fast resource extractors, but not both. Manipulating the C/P exchange rate by swapping the plant host genotype shifted this Pareto front, indicating that the exchange rate constrains possible AM fungal growth strategies. We show by mathematical modeling how AM fungal growth at fixed exchange rate leads to qualitatively different symbiotic outcomes depending on fungal traits and nutrient availability.

|
Scooped by
Jean-Michel Ané
February 12, 5:46 PM
|
Plants establish environmental connections through mycorrhizal symbiosis. These relationships enable them to obtain nutrients and cope with stress while simultaneously exchanging information through subterranean networks. A unified understanding of the molecular mechanisms underlying mycorrhizal interactions that drive adaptation and survival has not yet been achieved, in part because research on them stems from diverse fields of research, such as mycorrhizal ecology and plant epigenetics. This review presents recent studies demonstrating that epigenetic control serves as a central system enabling plants to adapt and maintain stable relationships with mycorrhizal fungi. We begin by describing different types of mycorrhizae. We then analyze mycorrhizal symbiosis by integrating plant and fungal genomic data with molecular evidence on DNA methylation, histone modification, chromatin remodeling, and small RNA pathways. We demonstrate that mycorrhizal symbiosis depends on changing chromatin states, which influence the regulation of the establishment, maintenance, and efficiency of symbiotic connections. They also regulate the balance between nutrient uptake and defense. They may underlie mycorrhizal stress and transgenerational “memory.” We review studies showing that RNA interference between different species enables reorganization of gene expression between plant and fungal cells. Finally, we identify key knowledge gaps and propose future research directions aimed at discovering reliable markers of mycorrhizal responses for epi-breeding and the development of climate-resilient agroecosystems.

|
Scooped by
Jean-Michel Ané
February 11, 4:53 PM
|
In this issue of Cell Host & Microbe, Hussain et al.1 demonstrate surprising microbiome resilience in oak trees under abiotic and biotic stress. In contrast to work on herbaceous plants or saplings, it was found that mature oak trees growing in situ experienced little microbiome change under stress, across both time and tissue type.

|
Scooped by
Jean-Michel Ané
February 10, 2:26 PM
|
Flavonoids, produced by the plant under nutrient stress, are required to initiate the legume-rhizobia symbiosis through the activation of rhizobial nod genes. Notwithstanding the central role of flavonoids in nodulation, their transcriptional regulation remains poorly understood. Here, we show that the nodulation signaling pathway 2 (NSP2) is required for transcriptional activation of flavonoid biosynthesis genes during nodulation in Medicago truncatula. Furthermore, MYB40, a legume-specific MYB transcription factor, is induced by rhizobia in the root epidermis. MYB40 directly binds to flavonoid biosynthetic gene promoters and is required for normal levels of nodulation. Biochemical and genetic evidence reveal that NSP2, not NSP1, interacts with MYB40 during rhizobial infection to strongly upregulate the symbiotic gene chalcone O-methyltransferase 1 in a manner dependent on MYB40 binding sites. Moreover, the overexpression of MYB40 and a microRNA-resistant NSP2 variant enhances nodulation under suboptimal rhizobial availability, suggesting this module fine-tunes symbiosis efficiency. Additionally, flavonoid regulation by NSP2 and MYB40 appears to facilitate arbuscular mycorrhizal colonization under nutrient starvation. Together, our findings establish an NSP2-MYB40 module that integrates symbiotic signaling with metabolic reprogramming, representing an evolutionary innovation for optimizing nitrogen acquisition in dynamic environments.

|
Scooped by
Jean-Michel Ané
February 6, 6:38 PM
|
Nitrogen is a major limiting nutrient for plant growth and a central driver of fertilizer use in agriculture. In many agricultural soils, nitrate is the main form of available nitrogen, but it is highly mobile and unevenly distributed, forcing plants to continuously adjust root growth to local supply. Legumes add an extra layer of complexity because they can also obtain ammonium through symbiotic nitrogen fixation in root nodules, which are carbon-expensive organs that must be regulated according to both external nitrate levels and internal nitrogen status. This review compares nitrogen signalling in the non-legume Arabidopsis thaliana and the model legume Medicago truncatula. Our aim is to understand how conserved pathways have been rewired to support these different nitrogen acquisition strategies. We focus on three core modules: CLE–CLV1/SUNN signalling, the NIN–NLP transcriptional module and the miR2111–TML pathway. In Arabidopsis, CLE–CLV1 acts mainly as a local low-nitrogen checkpoint that restricts lateral root emergence in nitrate-poor zones, while NLP7 integrates nitrate availability with root architectural responses. In Medicago, orthologous components have been recruited into the autoregulation of nodulation (AON) pathway, where nitrate-induced CLE peptides, NIN–NLP interactions and the systemic miR2111–TML module together integrate nitrate supply and rhizobial infection to coordinate nodulation with whole-plant nitrogen status and carbon cost. Within this network, the CLV1 ortholog SUNN acts as a central integration point. It responds to rhizobia-induced CLE peptides and to nitrogen-dependent systemic signals, including nitrate-induced CLE peptides, that influence lateral root growth in the absence of symbiosis. By contrasting these modules in Arabidopsis and Medicago, we show how legumes have layered symbiotic regulation onto ancestral nitrogen-signalling circuits that were first characterised in non-legume nitrate responses. This evolutionary perspective provides a mechanistic basis for future efforts to improve nitrogen use efficiency in crops by adjusting how CLE signalling, NIN/NLP activity and miR2111–TML-like modules link nitrogen status to root and nodule development.

|
Scooped by
Jean-Michel Ané
February 6, 6:22 PM
|
Nitrogen-fixing root nodule symbiosis (RNS) occurs in some eudicots, including legumes, and is regulated by the transcription factor NODULE INCEPTION (NIN), derived from the NIN-LIKE PROTEIN (NLP) family. However, how the NIN protein acquired RNS-specific functions remains unclear. We identify a previously undescribed motif in Lotus japonicus NIN, located downstream of the RWP-RK domain, which we term the FR. This motif broadens NIN’s DNA binding specificity by stabilizing the RWP-RK dimer interface. nin mutants lacking the FR motif show defective nodulation and impaired nitrogen fixation. Arabidopsis NLP2 carries a NIN-type FR and shares key features with NIN. Furthermore, the NIN-type FR had already emerged before the divergence of gymnosperm and angiosperm lineages, suggesting that a specific molecular feature of NIN involved in RNS regulation was inherited from ancestral NLPs prior to the emergence of RNS.

|
Scooped by
Jean-Michel Ané
February 4, 3:48 PM
|
•
A single origin of the RNS regulatory network, followed by lineage-specific refinements
•
Reconstructing the stepwise evolution of the RNS gene regulatory network
•
Paleohexaploidy at ∼110 mya provided founder genes crucial for later GRN-RNS assembly
•
Modules for cell wall remodeling and kinase signaling enable symbiosome formation

|
Scooped by
Jean-Michel Ané
February 4, 3:14 PM
|
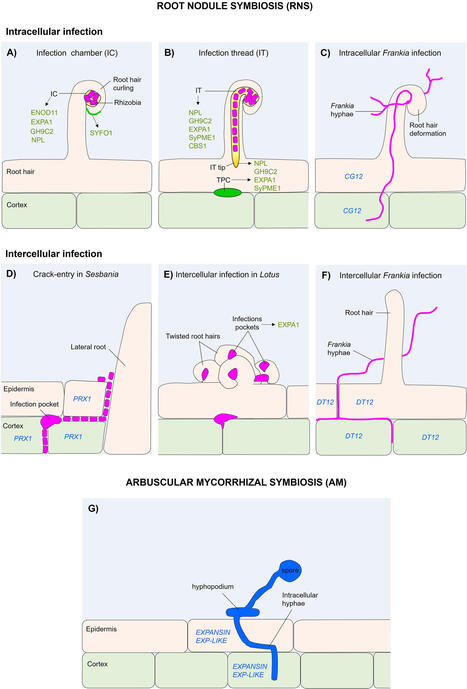
Plant roots are usually ground organs that perform essential roles, mostly associated with the anchoring of plants to the soil and absorption of nutrients and water. However, they are also exposed to a wide variety of microorganisms and may develop various symbiotic relationships, such as mutualism, which benefits both organisms. For instance, arbuscular mycorrhizal symbiosis is likely the oldest and most widespread mutualistic association, that occurs between plants and fungi. Another relevant example is the root nodule symbiosis, established between nitrogen-fixing bacteria and nodulating legumes, actinorhizal plants and Parasponia species. In both cases, microbial colonization of plant roots culminates in the formation of specialized symbiotic structures. In this regard, microbial infection is a critical step for the mutualistic relationship, where altering the cell wall biomechanics is necessary to facilitate microbial entry, which can be modulated by various cell wall protein families. This review examines the current knowledge on cell wall modifications occurring in plants roots during the symbiotic entry of microorganisms, focusing on the role of cell wall-remodeling proteins involved in these processes.

|
Scooped by
Jean-Michel Ané
January 31, 10:16 PM
|
Legume nodulation enables biological nitrogen fixation but is strongly repressed by nitrate. NIN-like proteins (NLPs) mediate this nitrate response, yet how their activity is regulated remains unclear. Here, we demonstrate that SUMOylation—a reversible posttranslational modification—is essential for the transcriptional activity and protein–protein interactions of MtNLP1 in Medicago truncatula, independently of its nitrate-induced nuclear localization. This modification is conserved in other NLPs, including Arabidopsis thaliana NLP7. Moreover, knockdown of SUMOylation-machinery components disrupts nodulation, suggesting that additional regulators in the symbiotic pathway also depend on SUMOylation. This work identifies SUMOylation as a conserved regulatory mechanism integrating nitrate signaling with root nodule symbiosis, with broad implications for improving plant nitrogen use efficiency.

|
Scooped by
Jean-Michel Ané
January 31, 9:04 PM
|
The cell surface localized lysin motif (LysM) receptor-like kinases/proteins (RLKs/RLPs) function as sensors for pathogenic and symbiotic microbes in land plants, perceiving chitin, lipo-chitooligosaccharides (LCOs), and peptidoglycan. LysM-RLKs/RLPs play a crucial role in activating various responses that lead to defense against pathogens or the establishment of symbiosis. While the functions of LysM-RLKs/RLPs were well-studied in land plants, their evolutionary origin and broader functional roles remain less explored. Streptophyte algae are widely recognized as the sister lineage of land plants. Land plants are believed to have emerged from a streptophyte algal ancestor. Plant–pathogen interactions are ancient and have played a pivotal role in shaping the evolution and complexity of the plant innate immune system. Genomic analyses revealed the presence of LysM-RLKs in two streptophyte algal species, Charophyceae Chara braunii and Zygnematophyceae Spirogyra pratensis. The functional roles of LysM-RLKs in both species were subsequently characterized. Through phylogenetic analysis combined with sequence alignment, I propose that LysM-RLKs originated from the last common ancestor of Charophyceae, Zygnematophyceae, and land plants. A conserved CXC motif within the LysM ectodomains (ECDs) was identified in streptophyte algal LysM-RLKs, like those present in land plants. Structural predictions using AlphaFold2 and three-dimensional modeling revealed that the overall architecture of LysM ECD is conserved, including a chitin-binding groove within the LysM2 domain. In vitro binding assays further demonstrated the chitin-binding capability of LysM ECDs from both streptophyte algae and bryophytes, suggesting an ancestral role of LysM ECDs in chitin recognition. Intriguingly, however, chitin treatment did not trigger downstream transcriptional responses in streptophyte algae, pointing to a functional divergence in LysM-RLKs during evolution. Furthermore, genetic and interaction studies demonstrated that heterodimerization and the formation of higher-order oligomeric complexes of LysM receptors are essential for proper function in bryophytes. In contrast to bryophytes, chitin treatment did not promote homodimerization of algal LysM receptors. Overall, this study new provides an insight into the evolutionary origin and functional diversification of LysM RLKs/RLPs in plants.
|

|
Scooped by
Jean-Michel Ané
February 16, 8:01 PM
|
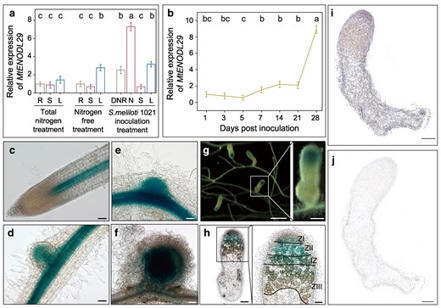
The early nodulin-like (ENODL) subfamily, part of the phytocyanin, arabinogalactan protein, and nodulin-like families, is involved in plant growth and stress resistance. However, its role in symbiotic nodulation remains poorly understood. In barrel medic (Medicago truncatula), we found MtENODL29 was strongly activated at the late stages of nodule development, particularly in the infection zone of nodules. Both RNA interference (RNAi) and mutation of MtENODL29 caused a considerable reduction in nodule numbers, an increase in cysteine protease activity, a dramatic decrease in leghemoglobin content, and signs of premature senescence in nodule cells, suggesting that disruption of MtENODL29 accelerates nodule aging. Transcriptome analysis of 7-dpi (day post inoculation) inoculated roots and 28-dpi nodules in enodl29 mutants showed significant downregulation of symbiotic genes, accompanied by differential expression of genes associated with lipid metabolism and transport. MtENODL29 mutation also negatively impacted plant growth and development. MtENODL29 bound to MtnsLTP (nonspecific lipid transfer protein), MtKCR (very-long-chain 3-oxoacyl-CoA reductase), and MtSec61γ (gamma subunit of the translocase complex Sec61) through its ALR (arabinogalactan protein-like region) domain. MtENODL29 co-localized with these proteins in the plasma membrane and endoplasmic reticulum. Notably, MtnsLTP showed high expression in the nodules, similar to MtENODL29, while MtKCR and MtSec61γ were also highly expressed in the leaves and stems. These results suggest that MtENODL29 participates in membrane lipid modification and transport by interacting with MtnsLTP, MtKCR, and MtSec61γ, facilitating the formation of symbiosome membranes as alfalfa rhizobium (Sinorhizobium meliloti) strain 1021 are released into nodule cells. Moreover, MtENODL29 influences plant growth, highlighting its role in coordinating plant development and symbiosis.

|
Scooped by
Jean-Michel Ané
February 12, 8:12 PM
|
Molybdenum (Mo) nitrogenase is a two-component enzyme complex that catalyzes the reduction of dinitrogen to ammonia and protons to hydrogen gas. We have shown that electrons for dinitrogen reduction can be delivered photochemically to the catalytic MoFe protein component by cadmium sulfide (CdS) nanocrystals. In this study, we used electron paramagnetic resonance spectroscopy to measure the transient populations of catalytic intermediates. We fit the populations with a pre-steady-state kinetic model, which allowed us to distinguish between productive and non-productive reaction pathways and extract the rate constants for the reaction. Our results demonstrated that the rate of catalytic electron delivery into MoFe protein increased with the concentration of the sacrificial electron donor. This enabled electron delivery to exceed the rate of hydride protonation, a relaxation pathway that competes with N2 binding. Thus, managing the balance between electron transfer and hole transfer reactions is required to achieve a kinetic regime that favors N2 reduction.

|
Scooped by
Jean-Michel Ané
February 12, 5:57 PM
|
Rhizobial type-Ⅲ effectors (T3Es) contribute to establishing symbiotic interactions with legume host plants, alongside Nod factors. However, the functions of most rhizobial T3Es, as well as the regulatory and molecular mechanisms underlying their symbiotic effects on hosts, particularly in soybean, are poorly documented. Here, we characterize the function of the T3E Nodulation Outer Protein C (NopC) in the broad-host-range rhizobium Sinorhizobium fredii HH103 for promoting symbiosis in soybean. NopC genotype influences root nodulation across diverse host germplasm and this is further influenced by GmRAC1, encoding a ROP/RAC family GTPase in soybean. GmRAC1 physically interacts with NopC to subsequently induce the expression of the essential symbiotic genes GmNIN2a/2b and GmENOD40. Knock-down of GmNIN2a/2b results in NopC failing to promote symbiosis, and Gmrac1 mutants have fewer nodules than the wild type. NopC facilitates multiple infection stages whereas the requirement for GmRAC1 is pronounced for infection-thread progression and nodule-primordia initiation. Natural variation in the GmRAC1 promoter largely dictates the symbiotic contribution of NopC during symbiotic establishment, and elite GmRAC1 haplotypes with strong expression were artificially selected in soybean breeding. Transgenic over-expression level and elite GmRAC1 haplotypes increase plant height, 100-seed weight and soybean yield. GmRAC1 serves as a key regulator of NopC-mediated symbiosis promotion and offers translational potential for enhanced symbiotic nitrogen fixation in molecular breeding of soybean.

|
Scooped by
Jean-Michel Ané
February 12, 5:48 PM
|
In the mutualism between leguminous plants and rhizobia bacteria, rhizobia live inside root nodules, creating potential for host genes to shape the rhizobial selective environment. Many host genes that affect symbiosis have been identified; however, the extent to which these genes affect selection acting on rhizobia is unknown. In this study, we inoculated 18 Medicago truncatula symbiotic mutants (including mutants that alter Nodule Cysteine-Rich (NCR) peptide production, plant defence, and nodule number regulation) with a mixture of 86 Sinorhizobium meliloti strains. Most mutations resulted in reduced host benefits, but the effects on rhizobial benefit (i.e., relative strain fitness) varied widely, revealing widespread host-by-strain fitness interactions. Genome-wide association analyses identified variants on rhizobial replicons pSymA and pSymB as important in mediating strain fitness responses to host mutations. Whereas most top variants affected rhizobial fitness with one host mutation (limited effect variants), nine affected fitness across six or more host mutations. These pervasive variants occurred primarily on pSymA, the symbiotic replicon, and include fixL and some metabolic genes. In contrast to the limited effect variants, variants with pervasive positive effects on strain fitness when host genes were mutated tended to adversely affect fitness in wild-type hosts. Competition assays across Medicago genotypes confirmed a pervasive role for one candidate (malonyl-CoA synthase), and AlphaFold multimer modelling suggests that many rhizobial top candidates could interact with host NCR peptides. Our results reveal how host genetic mutations alter strain fitness, setting the stage for improving rhizobial inoculants and breeding legume hosts better adapted to multi-strain environments.

|
Scooped by
Jean-Michel Ané
February 12, 5:32 PM
|
Plants encounter a myriad of microorganisms that can invade with detrimental or beneficial outcomes. Taking a membrane-centric point of view, we discuss the cellular events underlying microbe-induced cell signaling, the navigation of microbes within plant tissues, and the molecular exchanges occurring at plant–microbe interfaces. We discuss the implications of individual membrane lipids and the emerging role of membrane biophysics in non-self and modified-self sensing. We highlight the common themes underlying the active role of membranes during plant interactions with viruses, bacteria, oomycetes, and fungi, and define exciting directions for future research.

|
Scooped by
Jean-Michel Ané
February 11, 4:53 PM
|
Global forest biomes face increasing stressors and disease outbreaks that threaten ecosystem health. Tree-associated microbiota are vital for tree resilience, yet their responses to biotic and abiotic stressors in mature trees remain poorly understood. Using an experimental woodland plot of 144 Quercus petraea trees subjected to drought (rain exclusion), nutrient stress (ringbarking), and biotic treatments (bacterial pathogens and beetle larvae) to simulate acute oak decline, we tracked microbial communities in leaf, stem, and root/rhizosphere tissues across four time points over 2 years. Oak trees hosted distinct microbial communities across tissue types, which remained largely stable under stress. Rain exclusion significantly altered microbiota composition, though these changes explained less than 1% of total variance. Actinobacteriota, linked to drought tolerance, increased in the root/rhizosphere of rain-excluded trees. These findings reveal a surprising resilience of oak-associated microbial communities to environmental and biotic disturbances, highlighting their potential role in forest ecosystem stability.

|
Scooped by
Jean-Michel Ané
February 10, 11:00 AM
|
Pesticides are widely distributed in soils1,2,3, yet their effects on soil biodiversity remain poorly understood4,5,6,7. Here we examined the effects of 63 pesticides on soil archaea, bacteria, fungi, protists, nematodes, arthropods and key functional gene groups across 373 sites spanning woodlands, grasslands and croplands in 26 European countries. Pesticide residues were detected in 70% of sites and emerged as the second strongest driver of soil biodiversity patterns after soil properties. Our analysis further revealed organism- and function-specific patterns, emphasizing complex and widespread non-target effects on soil biodiversity. Pesticides altered microbial functions, including phosphorus and nitrogen cycling, and suppressed beneficial taxa, including arbuscular mycorrhizal fungi and bacterivore nematodes. Our findings highlight the need to integrate functional and taxonomic characteristics into future risk assessment methodology to safeguard soil biodiversity, a cornerstone of ecosystem functioning.

|
Scooped by
Jean-Michel Ané
February 6, 6:36 PM
|
Arbuscular mycorrhizal (AM) fungi are known to enhance plant drought tolerance, but the physiological mechanism behind this benefit remains unclear. One explanation is that AM colonization improves root hydraulic conductance (Kr), thereby facilitating more efficient water uptake under soil drying, though this mechanism remains highly debated. Here, we measured Kr in tomato (Solanum lycopersicum L.) and pea (Pisum sativum L.) with and without AM using a noninvasive rehydration technique under soil drying, and this was complemented with the evaporative flux method under hydrated conditions. AM colonization was manipulated either through soil sterilization or by using nonmycorrhizal mutants, ensuring precise control of AM status. In both species, AM colonization had no positive impact on Kr under both well-hydrated and drought conditions. The finding suggests that the improved drought performance often observed in AM-colonized plants is not due to enhanced root water transport capacity. Instead, AM-induced benefits under drought may be mediated by other physiological adjustments.

|
Scooped by
Jean-Michel Ané
February 4, 3:52 PM
|
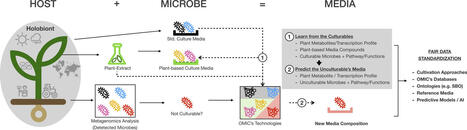
Background
The holobiont" refers to the plant and its associated microbiota that are pivotal to the plant's health, fitness, and survival. By in vitro culturing and functionally characterizing members of the plant microbiota, their specific roles in influencing plant responses to environmental changes can be determined and manipulated to foster sustainable agriculture and ecosystem management.
Aims
The review presents a comprehensive survey and current updates on culturomics of plant microbiota within the overall context of: a) the importance of understanding the plant holobiont composition and functioning; b) the necessity to in vitro track down and explore environmental microbiomes, entailing the plant microbiome with its myriad composition and spatio-temporal dynamics and mobility in various plant species, compartments and growth stages and c) the recent developments of the emerging in-situ similis cultivation strategies grounded on plant-based culture media.
Conclusions
The review highlights the urgent need to explore in vitro cultivation strategies built on compatible plant-based culture media, and the transformative role of omics technologies in refining these strategies. By bridging fundamental research and cultivation-based applications, such tools offer a gateway towards more sustainable and efficient in vitro cultivation systems, leading to a deeper understanding and potential manipulation of the plant holobiont.

|
Scooped by
Jean-Michel Ané
February 4, 3:37 PM
|
The Leguminosae (Fabaceae) is one of the largest and most evolutionary diverse plant families that comprises emblematic examples of symbiotic nitrogen fixation. This striking diversity is reflected across research areas ranging from taxonomy and phylogenomics to symbiosis, microbiology, biogeography, and functional traits. Insights into the genetic and molecular bases of root nodule symbiosis (RNS) are rapidly accumulating, enabling advances in agriculture and the engineering of biological nitrogen fixation (BNF). Nevertheless, substantial gaps remain, particularly regarding non-model species and the ecological and evolutionary drivers of RNS. This review summarises current knowledge on legume RNS and highlights that while the trait has likely contributed to legume diversification, the evolutionary trajectory of nodulation is complex, involving multiple gains, losses, and variations in symbiotic strategies across lineages. Nodule morphology and organogenesis, including determinate and indeterminate types, reveal structural and functional differences that may influence adaptability and BNF efficiency, although direct comparisons under varying environmental conditions remain limited. Ecological traits, such as drought tolerance, seed dormancy, and specialised pollination and defence mechanisms, interact with RNS to facilitate survival across diverse habitats. Case studies on Lebeckia ambigua and soybean wild relatives demonstrate how insights from non-model legumes can contribute to sustainable agriculture by improving stress resilience, expanding symbiotic partnerships, and broadening the genetic base for crop improvement. Future research should expand to non-model species and systematically assess nodulation, symbiotic efficiency, and environmental responsiveness to fully harness the potential of legumes for ecological and agricultural applications.

|
Scooped by
Jean-Michel Ané
February 2, 9:03 PM
|
Agriculture is under pressure to provide food for a growing population and the feedstock required to drive the bioeconomy. Methods to breed and genetically modify plants are inadequate to keep pace. When engineering crops, traits are painstakingly introduced into plants one-at-a-time, combine unpredictably, and are continuously expressed. Synthetic biology is changing these paradigms with new genome construction tools, computer aided design (CAD), and artificial intelligence (AI). “Smart plants” contain circuits that respond to environmental change, alter morphology, or respond to threats. Further, the plant and associated microbes (fungi, bacteria, archaea) are now being viewed by genetic engineers as a holistic system. Historically, plant health has been enhanced by many natural and laboratory-evolved soil microbes marketed to enhance growth or provide nutrients, or pest/stress resistance. Synthetic biology has expanded the number of species that can be engineered, increased the complexity of engineered functions, controlled environmental release, and can assemble stable consortia. New CAD tools will manage genetic engineering projects spanning multiple plant genomes (nucleus, chloroplast, mitochondrion) and the thousands of genomes of associated bacteria/fungi. This review covers advanced genetic engineering techniques to drive the next agricultural revolution, as well as push plant engineering into new realms for manufacturing, infrastructure, sensing, and remediation.

|
Scooped by
Jean-Michel Ané
January 31, 10:08 PM
|
Flavonoids are major plant secondary metabolites that mediate diverse plant–microbe interactions, including ectomycorrhizal (ECM) symbioses. However, their regulatory roles during ECM development remain poorly understood. Here, we investigated whether inoculation with Suillus bovinus alters flavonoid biosynthesis in Pinus yunnanensis roots and assessed how these flavonoids on fungal growth and gene expression. We applied exogenous flavonoids to S. bovinus mycelia to investigate fungal transcriptional and metabolic responses. Following inoculation, differentially expressed genes in P. yunnanensis roots were significantly enriched in the flavonoid biosynthesis pathway. Key enzyme-coding genes, including PAL, CHS, CHI, F3H, and FLS, were upregulated, and this was associated with increased flavonoid accumulation and enhanced antioxidant capacity. In S. bovinus, exogenous flavonoids promoted mycelial growth and induced metabolic adjustments related to carbohydrate and amino acid utilization. Several small secreted protein-related genes showed transcriptional responses to flavonoid exposure, indicating potential transcriptional modulation, although their specific roles in symbiosis remain unclear. These findings indicate that flavonoids may contribute to reciprocal interactions between host roots and ECM fungi and provide a molecular basis for further investigation.
|





 Your new post is loading...
Your new post is loading...