Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
November 16, 11:38 AM
|
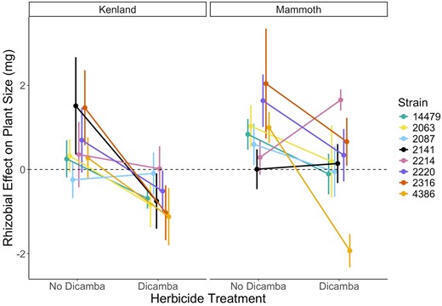
Agriculture has intensified the presence of chemical stressors in the rhizosphere—the region surrounding roots where critical plant-microbe interactions occur, such as those between leguminous plants and nitrogen-fixing rhizobial bacteria. Particularly, rhizospheric pesticide exposure can disrupt the efficacy of the plant-rhizobia mutualism and reduce plant productivity. However, it is unknown whether genetic variation in plants (GP), rhizobia (GR), or interactions between them and the pesticide environment (E), i.e., GP or R ×E, or GP ×GR ×E, could mitigate these negative outcomes. We grew two genotypes of the leguminous plant Trifolium pratense in symbiosis with each of eight genetic strains of its rhizobial partner Rhizobium spp. symbiovar trifolii. We exposed symbionts to the contemporary synthetic auxin herbicide dicamba or a control in the rhizosphere, and evaluated the symbiotic interaction and plant growth. Our results provide new evidence that rhizobial genetic variation drives herbicide impacts on mutualism outcomes through GR × E interactions. Rhizospheric herbicide delayed rhizobial colonization of plants via root nodule formation, but its effects on the number of nodules and fixed nitrogen produced varied depending on rhizobial strain. Similarly, while herbicide exposure reduced plant size on average, the degree of this effect was mediated by rhizobial partner, suggesting that rhizobia could potentially function as an “extended genotype” for defense against herbicide damage. As the use of herbicides, particularly synthetic auxins, continues to escalate, our findings have important implications for how certain rhizobia could be selected to improve plant fitness in the face of these anthropogenically-released chemicals.

|
Scooped by
Jean-Michel Ané
November 16, 11:10 AM
|
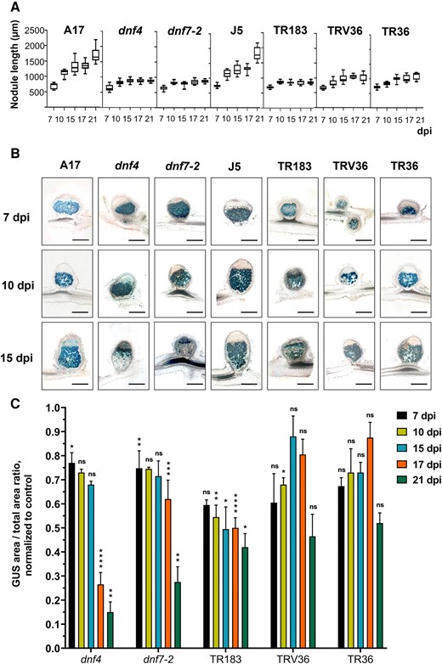
Nodule senescence in barrel medic (Medicago truncatula) can occur as a natural, developmentally regulated process or be triggered prematurely by environmental stress or ineffective symbiotic interactions. In this study, we examined five M. truncatula Fix⁻ mutants (dnf4, dnf7-2, TR183, TRV36, and TR36) that fail to fix nitrogen to determine whether they share common senescence-related traits. Our findings reveal that, despite distinct genetic defects, all mutants exhibit similar hallmarks of premature senescence: a rapid decline in the transcription of nitrogen-fixation-related genes (as indicated by DINITROGENASE REDUCTASE (NifH) expression), early degradation of bacteroids and symbiotic cells, recolonization of nodules by saprophytic rhizobia, premature closure of the nodule endodermis, impaired postmitotic differentiation of the symbiotic cells, and upregulation of senescence marker genes (CYSTEINE PROTEASE 2 (CP2), CYSTEINE PROTEASE 6 (CP6), CHITINASE 2, and PURPLE ACID PHOSPHATASE 22 (PAP22). Neither symbiotic maintenance genes (DEFECTIVE IN NITROGEN FIXATION 2 (DNF2), Symbiotic CYSTEINE-RICH RECEPTOR-LIKE KINASE (SymCRK), and REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD)) that inhibit plant defense responses nor the defense-related gene PATHOGENESIS-RELATED PROTEIN 10.1 (PR10.1) were upregulated, suggesting that premature senescence in these mutants is driven primarily by proteolytic activities rather than immune responses. These results indicate that early nodule senescence is a common feature of ineffective M. truncatula–Sinorhizobium medicae interactions, independent of the specific genetic mutation. Understanding nodule longevity and functionality may contribute to the development of strategies to enhance symbiotic efficiency in legumes for sustainable agriculture.

|
Scooped by
Jean-Michel Ané
November 15, 12:47 PM
|
•Peanut genetic diversity influenced AMF and diazotrophic community structures.
•Peanut diversity increased AMF α-diversity but reduced diazotrophic α-diversity.
•Peanut diversity decreased AMF dispersion but increased diazotrophic dispersion.
•Peanut genetic diversity increased AMF network complexity and stability
•Peanut genetic diversity lowered network complexity and stability of diazotrophs.

|
Scooped by
Jean-Michel Ané
November 15, 12:36 PM
|
Plants require fixed nitrogen, which is often the growth-limiting nutrient. Legume plants such as beans, lentils and soybeans can overcome a shortage of nitrogen by forming a root nodule endosymbiosis with nitrogen-fixing rhizobium bacteria. These bacteria possess the biochemical machinery to convert atmospheric dinitrogen gas (N2) into ammonia (NH4+), which they exchange with the plant in return for carbon sources and other essential nutrients. Legumes can fix up to 400 kg of nitrogen per hectare, making the rhizobium nodule endosymbiosis an appealing alternative to environmentally harmful chemical nitrogen fertilizers

|
Scooped by
Jean-Michel Ané
November 14, 10:14 AM
|
This book explores harnessing mycorrhiza in hilly agriculture and presents innovative strategies for sustainable cultivation. Exploring the symbiotic relationship between plants and mycorrhizal fungi reveals how this partnership can optimize soil health, enhance nutrient uptake, and increase crop resilience in challenging terrains. From understanding the ecological role of mycorrhiza to practical applications in hilly regions, the book highlights the use of mycorrhiza in hill agriculture, including details on mycorrhizal diversity, its impact on hill crops, and how to maintain and promote mycorrhizal diversity for better sustainability and economic viability. By addressing the unique challenges faced by hillside farmers, it aims to improve agricultural practices, mitigate erosion, and promote long-term environmental sustainability.
This book is essential for policymakers seeking sustainable agricultural strategies, researchers exploring beneficial fungi, soil scientists, agronomists, progressive farmers and students pursuing agriculture sciences.

|
Scooped by
Jean-Michel Ané
November 12, 2:27 PM
|
•Microplastics increased glomalin but had limited effects on microbial necromass.
•Microplastics significantly altered arbuscular mycorrhizal fungi (AMF) diversity.
•Microplastics improved SOC sequestration through changes in AMF and glomalin.
•Biodegradable microplastics showed greater benefits for soil carbon stabilization.

|
Scooped by
Jean-Michel Ané
November 9, 10:18 PM
|
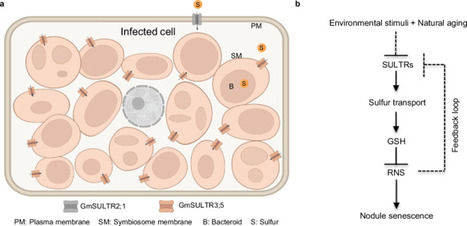
Symbiotic nitrogen fixation (SNF) in legume-rhizobia represents a sustainable and eco-friendly alternative to chemical nitrogen fertilizers in agriculture. Identifying key factors involved in nodule senescence, is crucial for enhancing SNF by effectively extending the lifespan of nodules. Here, we reveal that sulfur (S), an essential element for SNF, plays a major regulatory role in the senescence of soybean (Glycine max) nodules. Blocking S input into the symbiosome by knocking out either S transporter genes SULTR2;1 or SULTR3;5, resulted in a significant decrease in glutathione levels. This reduction impairs the capacity for reactive nitrogen species scavenging, thereby accelerating nodule senescence. Notably, reducing reactive nitrogen species (RNS) production in rhizobia or increasing S input in soybean nodules through genetic manipulation, can effectively mitigate high nitrogen-induced nodule senescence. Our findings demonstrate that SULTR-mediated S input is a pivotal step in regulating nodule senescence, and provide insights for developing strategies to enhance SNF in legumes by delaying nodule senescence.

|
Scooped by
Jean-Michel Ané
November 9, 12:46 PM
|
Plants and microbial organisms develop close symbioses that have a significant influence on agricultural productivity and plant health. These 'agricultural engines' have continuously supported balancing global food security from historical times. Against the backdrop of the global challenges faced by modern agriculture, including soil degradation and over-reliance on synthetic inputs, this review examines the intricate relationships within the soil microbiome, and their impact on sustainable crop production. It further investigates the pivotal functions of these partnerships in nutrient cycling, biotic stress suppression, hormone modulation, and stimulating and enhancing the flourishing growth of crops. Highlighting the importance of plant-microbe relationships, this study explores the potential of biological nitrification inhibitors, biocontrol agents, and biofertilizers, specifically, nitrogen-fixing bacteria and phosphorus-solubilizing microbes, to optimize nutrient use efficiency, suppress biotic stress, enhance nutrient availability for crops, and mitigate climate change. Furthermore, challenges related to environmental factors and the commercial adoption of microbial products are also scrutinized. The review concludes by outlining future research directions and envisioning the integration of microbial partnerships into sustainable climate resilience agricultural practices, thereby offering a holistic approach to address current agricultural challenges and pave the way for a more resilient and environmentally friendly food production system. This will help guide cutting-edge microbiome-based solutions, to improve global food production and agricultural resource use efficiency in the years to come.

|
Scooped by
Jean-Michel Ané
November 7, 3:59 PM
|
• Legume symbiosis with rhizobial nitrogen-fixing bacteria enable them to grow in nitrate-depleted soils. Rhizobial symbioses also induces systemic plant defence against bioagressors. • We investigated how nitrogen-fixing symbiosis (NFS) in the legume Medicago truncatula can prime plant defence against the pea aphid Acyrthosiphon pisum. We analysed metabolite modification both by gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) and defence pathway gene expression by qPCR in leaves of both NFS and nitrate-fed (non-inoculated; NI) plants after aphid infestation (Amp). • The accumulation of primary and secondary metabolites was modulated by both NFS and aphid infestation. Sixty two defense-related metabolites such as salicylate, pipecolate, gentisic acid and several soluble sugars were differentially regulated by aphid infestation in both NFS and NI conditions. Nineteen metabolites, including triterpenoid saponins, accumulated specifically in NFS_Amp conditions. Gene expression analysis showed that aphid-infested plants exhibited significantly higher expression of Chalcone isomerase, flavonol synthase, hydroxyisoflavone-O-methyl transferase and Pterocarpan synthase, while D-pinitol dehydrogenase was only significantly induced in NI-infested leaves. • Our data suggest that NFS, in addition to being a plant nitrogen provider, stimulates specific legume defenses upon pest attack and should also be considered as a potential tool in Integrated Pest Management strategies.

|
Scooped by
Jean-Michel Ané
November 7, 3:53 PM
|
Plant cells constantly monitor both their surroundings and internal state, enabling dynamic responses to developmental and environmental cues. Central to this sensing capacity are membrane-bound receptor kinases (RKs), one of the largest and most functionally diverse protein families in plants. RKs detect a broad spectrum of signals, including microbial molecules, endogenous ligands, and changes in cellular status. They share a conserved modular architecture: an extracellular domain for signal perception, a single transmembrane domain, and a conserved cytoplasmic kinase domain that initiates intracellular signalling. Although structurally analogous to their animal counterparts, plant RKs evolved independently [1,2]. Their functional diversity stems primarily from their extracellular domains, with over 20 distinct structural classes described to date [1–3]. The repertoire of RKs expressed in a given cell type varies, as some RKs are ubiquitously expressed across tissues, while others are restricted to specific organs or developmental contexts [4].

|
Scooped by
Jean-Michel Ané
November 7, 3:45 PM
|
The extensive use of synthetic nitrogen fertilizers has sustained global food production for more than a century but at high environmental and energetic costs. Improving ni-trogen use efficiency (NUE) has therefore become a key objective to maintain produc-tivity while reducing the ecological footprint of agriculture. This review synthesizes current knowledge on the biological foundations of NUE enhancement, focusing on the role of microbial biofertilizers and biostimulants. The main mechanisms through which plant-associated microorganisms contribute to nitrogen acquisition and assimi-lation are analyzed. In parallel, advances in genomics, biotechnology, and formulation science are highlighted as major drivers for the development of next-generation mi-crobial consortia and bio-based products. Particular attention is given to the current landscape of commercial biofertilizers and biostimulants, summarizing the principal nitrogen-fixing and plant growth–promoting products available on the market and their agronomic performance. Moreover, major implementation challenges are dis-cussed, including formulation stability and variability in field results. Overall, this re-view provides an integrated perspective on how biological innovations, market evolu-tion, and agronomic optimization can jointly contribute to more sustainable nitrogen management and reduce dependence on synthetic fertilizers in modern agriculture.

|
Scooped by
Jean-Michel Ané
November 6, 9:53 AM
|
Bacterial genome evolution is characterized by gains, losses, and rearrangements of functional genetic segments. The extent to which large-scale genomic alterations influence genotype-phenotype relationships has not been investigated in a high-throughput manner. In the symbiotic soil bacterium Sinorhizobium meliloti, the genome is composed of a chromosome and two large extrachromosomal replicons (pSymA and pSymB, which together constitute 45% of the genome). Massively parallel transposon insertion sequencing (Tn-seq) was employed to evaluate the contributions of chromosomal genes to growth fitness in both the presence and absence of these extrachromosomal replicons. Ten percent of chromosomal genes from diverse functional categories are shown to genetically interact with pSymA and pSymB. These results demonstrate the pervasive robustness provided by the extrachromosomal replicons, which is further supported by constraint-based metabolic modeling. A comprehensive picture of core S. meliloti metabolism was generated through a Tn-seq-guided in silico metabolic network reconstruction, producing a core network encompassing 726 genes. This integrated approach facilitated functional assignments for previously uncharacterized genes, while also revealing that Tn-seq alone missed over a quarter of wild-type metabolism. This work highlights the many functional dependencies and epistatic relationships that may arise between bacterial replicons and across a genome, while also demonstrating how Tn-seq and metabolic modeling can be used together to yield insights not obtainable by either method alone.

|
Scooped by
Jean-Michel Ané
November 5, 4:31 PM
|
WrtF is a glycosyltransferase that incorporates fucose residues into O-polysaccharide, resulting in a relatively hydrophobic O-polysaccharide that helps host plants recognize these bacteria as commensals. This enzyme belongs to a different fold family than previously characterized fucosyltransferases and, unusually, operates without assistance from either metal cations or positively charged amino acid side chains. We also show that the protein stably binds GDP-fucose in a conformation that is incompatible with acceptor binding and catalysis, implying that the enzyme must reposition the substrate prior to turnover. The observed binding mode is specific to fucose and may help minimize the misincorporation of off-target sugars. These findings expand our knowledge of the range of substrate recognition and catalytic strategies glycosyltransferases can use.
|

|
Scooped by
Jean-Michel Ané
November 16, 11:34 AM
|
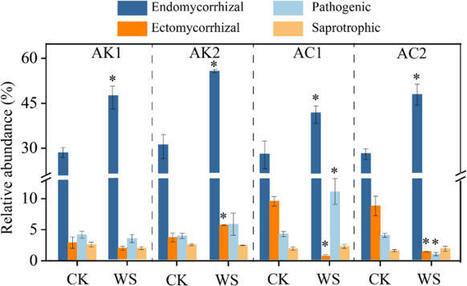
Purpose
Fungi play a crucial role as soil microorganisms, mediating carbon and nitrogen dynamics while influencing nutrient availability for host plants. However, the regulatory effects of plants on fungal communities and the functional roles of soil fungi in plant-soil system dynamics, as well as the nitrogen transformation rates in different pH soils remain largely uncharacterized.
Methods
In this study, wheat (Triticum aestivum L.), a typical nitrate-preferring crop, was selected as the model plant in alkaline (AK) and acidic (AC) soils. After one month pot cultivation, the wheat planted soil (WS) and not planted (CK) soil samples were collected. Fungal community structure and diversity in soil samples were assessed through high-throughput sequencing of the ITS1 region. Plant samples were dried and sieved for nitrogen concentration and uptake rate analysis. The soil and plant nitrogen transformation rates were quantified using 15N isotope techniques.
Results
We observed divergent responses of fungal community structure and diversity to wheat cultivation between AK and AC. In AC, wheat cultivation increased fungal copies and Ascomycita abundance, while in AK, fungal community structure and diversity shifts were contingent on DOC and nitrogen content. Notably, the cultivation of wheat in AK and AC soils significantly reduced the concentration of soil NH4⁺ and NO3⁻, and significantly promoted the uptake rate of NO3⁻. FUNGuild functional classification revealed a dominant of endomycorrhizal across all treatments. As the abundance of endomycorrhizal fungi increased with the decreased soil inorganic nitrogen. In contrast, ectomycorrhizal fungi abundance decreased. The abundance of endomycorrhizal fungi showed a strong positive correlation with the gross mineralization rate (P < 0.05) and the nitrification rate (P < 0.05).
Conclusions
Our findings suggest that wheat cultivation promotes the recruitment of endomycorrhizal fungi while stimulating soil nitrogen mineralization and nitrification processes, ultimately enhancing nitrogen uptake efficiency in wheat plants. Those findings reveal the critical role of mycorrhizal fungi in mediating wheat nitrogen acquisition, underscoring the necessity of integrating endomycorrhizal fungi into nitrogen transformation models to refine the accuracy of simulating plant-soil nitrogen turnover dynamics.

|
Scooped by
Jean-Michel Ané
November 15, 12:47 PM
|
Chickpea (Cicer arietinum L.) is the second most important food legume crop, capable of converting atmospheric nitrogen (N2) into ammonia (NH3) in symbiotic association with Mesorhizobium cicero through a process called biological nitrogen fixation (BNF). BNF shows promise in effectively diminishing reliance on exogenous nitrogen applications, enhancing soil sustainability and productivity in pulse crops. Notably, there are limited studies on the molecular basis of root nodulation in chickpea. In order to identify new sources of highly nodulating genotypes and gain deep insights into genomic regions governing BNF, a diverse chickpea global germplasm collection (284) was evaluated for nodulation and yield traits in four different environments in an augmented randomized block design. The genotypes exhibited significant trait variation, encompassing all traits under study. Correlation analysis revealed a significant positive correlation of nodulation traits on yield within the chickpea population. The genotypes ICC 7390, ICC 15, ICC 8348, and ICC 2474 were identified as high nodulating across the locations. Genome-wide association studies (GWAS) identified noteworthy and stable marker–trait associations (MTAs) linked to the traits of interest. For the traits number of nodules (NON) and nodule fresh weight (NFW), 65 and 109 significant MTAs were identified, respectively. In addition, two SNPs, Ca1pos289.52482.1 and 6_33340878, identified in our earlier studies were validated by independent population studies, which are crucial in evaluating the accuracy and reliability of the projections. Subsequent analysis revealed that a substantial proportion of these MTAs were situated within intergenic regions, with the potential to modulate genes associated with the focal traits. The candidate genes identified could be converted to Kompetitive allele-specific PCR (KASP) markers and exploited in marker-assisted breeding, accentuating their impact on future chickpea breeding efforts.

|
Scooped by
Jean-Michel Ané
November 15, 12:44 PM
|
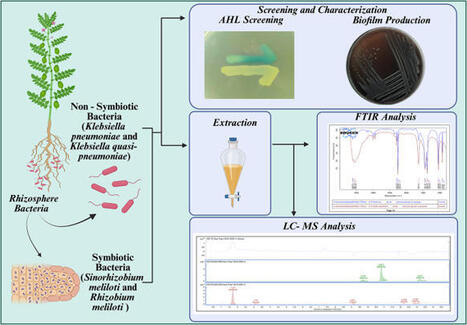
Quorum sensing (QS) is a critical bacterial communication system in the plant rhizosphere regulating interactions and biofilm formation, which are essential for survival. While AHL profiles in rhizobacteria are important for sustainable agriculture, a comprehensive understanding of their diversity, particularly in beneficial, non-hemolytic strains, remains limited. This study addresses this gap by providing a comparative analysis of N-acyl homoserine lactone (AHL) profiles and biofilm capabilities in a panel of symbiotic Ensifer meliloti and non-symbiotic Klebsiella pneumoniae and Klebsiella quasi-pneumoniae strains. For definitive taxonomic identification, the two MTCC strains (Ensifer meliloti RM and SINO) were subjected to 16S rRNA gene sequencing and phylogenetic analysis. Initial biosensor screening confirmed AHL production in all tested strains. Four strong AHL producers were selected for in-depth analysis. While Autoinducer Type-II (AI-2) signaling was absent, all strains demonstrated robust biofilm formation. Detailed LC–MS/MS profiling revealed distinct AHL profiles for each strain, highlighting significant intra-species diversity. Notably, this study reports the first identification of N-3-oxo-hexanoyl-HSL (C6-oxo-HSL) in Ensifer meliloti strain RM (10499). Furthermore, the non-hemolytic Klebsiella strains exhibited a profile dominated by 3-O-C8-HSL and 3-O-C12-HSL, which significantly differs from profiles reported in pathogenic Klebsiella strains, suggesting a link between QS systems and bacterial lifestyle. The identification of these specific AHL signals in robust biofilm-forming, non-hemolytic bacteria underscores their potential as safe and effective bioinoculants for enhancing rhizosphere colonization and supporting sustainable agriculture.

|
Scooped by
Jean-Michel Ané
November 14, 10:19 AM
|
The paradigms of legume-rhizobia symbiosis are derived primarily from conserved features of Inverted- Repeat Lacking Clade (IRLC) legumes and closely related species. The Dalbergioids diverged from the IRLC early in legume evolution and possess unique symbiotic features but few genetically tractable models. The small, diploid Dalbergioid Aeschynomene americana (American jointvetch) has promise as a genetic model for Dalbergioid-rhizobia symbiosis, yet only a few studies have examined its symbiotic properties.
We examined the symbiont range of A. americana from central Florida and characterized a native A. americana nodule isolate, Bradyrhizobium sp. USDA3516.
We find that A. americana forms effective symbioses with B. sp. USDA3516, which is closely related to Thai A. americana symbiont B. sp DOA9, and with symbionts from the Dalbergioids stylo and peanut. Interestingly, several strains that effectively nodulated A. americana exhibited branched bacteroid morphologies, but we found that branching was neither necessary nor sufficient for effective symbiosis.
Our study contradicts the prevailing view that bacteroid shape is a major determinant of symbiotic efficiency and presents the A. americana-B. sp. USDA3516 interaction as an optimal model of A. americana symbiosis.

|
Scooped by
Jean-Michel Ané
November 14, 10:12 AM
|
Soybean is valued for its high protein content and its symbiosis with the nitrogen-fixing bacterium Bradyrhizobium japonicum. This study evaluated whether commercial inoculants could mitigate drought effects in soybean grown in pots at 20% (drought) and 70% (control) field water capacity. Inoculation with Rhizobium Bio-Gen improved photosystem II efficiency under water deficit, while Nitragina IUNG prevented drought-induced losses in water status, biomass, nodulation, and yield. Nitragina Biofood promoted the highest share of biologically fixed nitrogen under optimal watering (leaves: 65.5%; seeds: 57.0%) and further enhanced N2 fixation during drought (leaves: 79.9%; seeds: 57.8%). Across watering regimes, δ13C values were highest in leaves and lowest in pods and seeds, indicating drought-driven recycling of respired CO2. Overall, application of Nitragina IUNG in soybean cultivation can effectively mitigate the adverse effects of soil drought on yield, whereas Nitragina Biofood appears particularly well suited for crops intended as green manure or as preceding crops for winter cereals.

|
Scooped by
Jean-Michel Ané
November 10, 5:49 PM
|
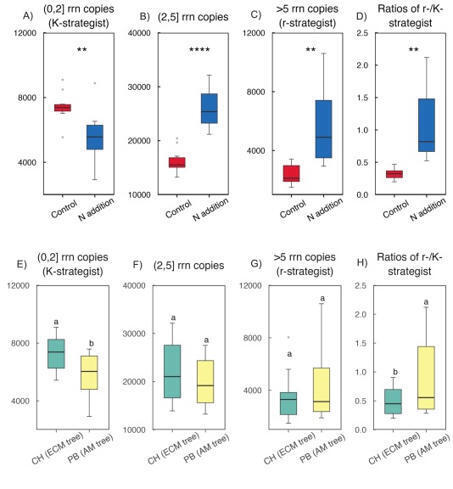
Plants establish symbiotic relationships with various mycorrhizal fungi, which may represent a crucial mechanism for different modes of nutrient cycling and soil ecological processes. However, our understanding of rhizosphere-specific microbial traits—such as fungal functional guilds and bacterial life-history strategies (copiotrophic vs. oligotrophic)—at the individual mycorrhizal tree species level remains limited. In this study, we examined how N addition (47.5 g N m−2 yr−1) affects bacterial and fungal communities in the rhizosphere of two dominant subtropical tree species: Castanopsis hystrix, an ectomycorrhizal (ECM) tree species, and Phoebe bournei, an arbuscular mycorrhizal (AM) tree species. We used amplicon sequencing and ecological trait-based analyses to conduct this research. Nitrogen enrichment led to a reduction in bacterial α-diversity, favouring copiotrophic Gammaproteobacteria and r-strategists while suppressing oligotrophic groups, such as Acidobacteriia, Alphaproteobacteria, and Acidimicrobiia, along with K-strategists. Furthermore, adding N increased heterogeneity in bacterial β-diversity between mycorrhizal plant types, resulting in divergent shifts in copiotrophic and oligotrophic bacterial groups. This shift amplified community differentiation specific to the mycorrhizal plant type rather than promoting convergence. Fungal responses to N addition varied based on the host mycorrhizal plant type. In AM-associated P. bournei, N addition decreased the relative abundance of symbiotrophic AM fungi and reduced fungal α-diversity. Conversely, in ECM-associated C. hystrix, N addition suppressed both saprotrophic and symbiotrophic ECM fungi while increasing α-diversity, likely due to the growth of pathotrophic taxa. Despite these contrasting responses, N addition homogenised fungal β-diversity across mycorrhizal plant types, reducing differences among mycorrhizal-specific fungal guilds. Structural equation modelling revealed that soil N and P availability were the primary drivers of bacterial community restructuring. In contrast, fungal assemblages were impacted by both soil chemistry and root traits, notably fine root length. These findings highlight that N enrichment disrupts mycorrhizal plant type-specific microbial niche partitioning in subtropical forests, favouring copiotrophic bacteria and separating fungal communities from host identities, a potential mechanism driving ecosystem-level functional changes under elevated N deposition.

|
Scooped by
Jean-Michel Ané
November 9, 5:34 PM
|
An exquisite symbiotic relationship between legumes and rhizobia leads to the development of nitrogen-fixing special organelles known as nodules in nitrate-deficient environments, whereas a high level of nitrate in soil negatively regulates the pleiotropic phases of root nodule symbiosis (RNS), including rhizobial infection, nodule organogenesis and leghemoglobin synthesis. Here, we identified a special group of nodule-specific non-symbiotic leghemoglobin genes (AhLghs) in the crack entry legume peanut; however, their functional role and transcriptional regulation remain enigmatic. A comparative transcriptomic analysis revealed that the downregulation of nodule inception (AhNIN) and non-symbiotic leghemoglobin (AhLghs) genes played a pivotal role in the nitrate-mediated inhibition of nodulation in peanut. The knockdown of AhLghs and overexpression of AhLgh1 resulted in lower and higher leghemoglobin content, respectively, corroborating their roles as positive regulators of nitrogen fixation in peanut. On the other hand, knockdown of AhNINs not only inhibited root nodulation but also decreased leghemoglobin content in peanut. Further, the DNA-affinity purification sequencing (DAP-Seq) analysis identified various nodulation genes, including AhLghs, as targets of AhNINs. After validating DNA-protein interaction by EMSA, the transactivation assay revealed that AhNINs can positively regulate AhLgh1 after binding to the NIN RESPONSIVE CIS ELEMENT (NRCE) of its promoter. Our work bridges a critical gap in understanding how nitrate influences non-symbiotic leghemoglobin expression by targeting rhizobia-induced NINs in peanut, and offers a potential model suggesting that the nitrate-NIN-Lgh module might represent a key evolutionary event in fine-tuning root nodulation.

|
Scooped by
Jean-Michel Ané
November 7, 4:01 PM
|
Arbuscular mycorrhizal (AM) symbiosis develops through successive colonization of root epidermal and cortical cells, culminating in the formation of arbuscules, tree-like intracellular structures that are transient yet essential sites of nutrient exchange. To dissect the cellular and structural complexity of AM establishment in rice roots colonized by Rhizophagus irregularis, we applied dual-species spatial transcriptomics to simultaneously monitor plant and fungal gene transcripts at single-cell resolution. This approach revealed surprising differences in transcriptional activity between fungal structures and showed that morphologically similar arbuscules can be transcriptionally distinct. These findings suggest hidden functional diversity among arbuscules at single-cell resolution. Because arbuscules form and degenerate within only a few days, we further sought to capture translational activities across their life span. We pioneered AM-inducible TRAP-seq (Translating Ribosome Affinity Purification followed by RNA-seq) using stage-specific promoters, enabling cell-type- and stage-resolved profiling in AM symbiosis. This revealed extensive spatiotemporal reprogramming of nutrient transport and signalling, with distinct sets of phosphate, nitrogen, and carbon transporters and regulators induced or repressed at different stages of arbuscule development, suggesting that nutrient exchange is dynamically regulated across the arbuscule life cycle. More broadly, cell wall biosynthesis genes and key defence markers were suppressed during arbuscule formation, whereas at a later stage, defence markers were strongly upregulated, suggesting a host-driven shift towards arbuscule termination. Together, these findings highlight the nuanced and dynamic regulation of AM symbiosis at the cellular level, refining our understanding of how nutrient exchange and fungal development are coordinated in space and time.

|
Scooped by
Jean-Michel Ané
November 7, 3:57 PM
|
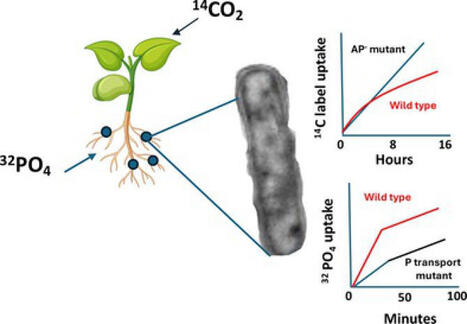
Bacteroid inorganic phosphorus (Pi) metabolism in the Rhizobium‐legume symbiosis differs between indeterminate and determinate legume nodules. In contrast to alfalfa bacteroids, bean ( Phaseolus vulgaris ) bacteroids exhibit high levels of alkaline phosphatase (AP), the native reporter enzyme for the bacterial Pi stress response. 14C and 32Pi whole plant labelling techniques were used in conjunction with diagnostic mutants (lacking AP or lacking high affinity Pi transport) to assess the relative importance of the Pi stress response in Rhizobium tropici bacteroids during symbiosis. The AP‐ mutant was not defective for symbiosis and did not differ from wildtype bacteroids for Pi acquisition. 14C‐CO2 feeding to host plants revealed 14C‐carbon uptake and accumulation in AP‐ mutant bacteroids, and their nodules were increased relative to wildtype bacteroids, implying that organo‐P compounds may account for meaningful levels of carbon exchange between symbionts. 32Pi tracer experiments implied that the high affinity transporter is important to bacteroid Pi acquisition and symbiotic performance in determinate nodules, but that the symbiosome Pi concentration does not meet the capacity of the high affinity transporter. 32P tracer work also illustrated that Pi taken up into the nodule does not remain in the nodule, but rather is redistributed to the host.

|
Scooped by
Jean-Michel Ané
November 7, 3:49 PM
|
The CLAVATA signaling pathway regulates plant development and plant–environment interactions. CLAVATA signaling consists of mobile, cell-type or environment-specific CLAVATA3/ESR-related (CLE) peptides, which are perceived by a receptor complex consisting of leucine-rich repeat receptor-like kinases such as CLAVATA1 and receptor-like proteins such as CLAVATA2, which often functions with the pseudokinase CORYNE (CRN). CLAVATA signaling has been extensively studied in various plant species for its developmental role in meristem maintenance. In addition, CLAVATA signaling was implicated in plant–microbe interactions, including root nodule symbiosis and plant interactions with mutualistic arbuscular mycorrhizal (AM) fungi. However, knowledge on AM symbiosis regulation by CLAVATA signaling is limited. Here, we report a dual role for Medicago truncatula CRN in development and plant–microbe interactions. In shoots, MtCRN modulates inflorescence meristem branching. In roots, the MtCRN promoter is active in vascular tissues and meristematic regions. In addition, MtCRN expression is activated in cortex cells colonized by AM fungi and negatively regulates root interactions with these microbes in a nitrogen-dependent manner; negative AM symbiosis regulation by CRN was also observed in the monocot Zea mays, suggesting this function is conserved across plant clades. We further report that MtCRN functions partially independently of the AM autoregulation signal MtCLE53. Transcriptomic data revealed that M. truncatula crn roots display signs of perturbed nutrient, symbiosis, and stress signaling, suggesting that MtCRN plays various roles in plant development and interactions with the environment.

|
Scooped by
Jean-Michel Ané
November 6, 11:40 AM
|
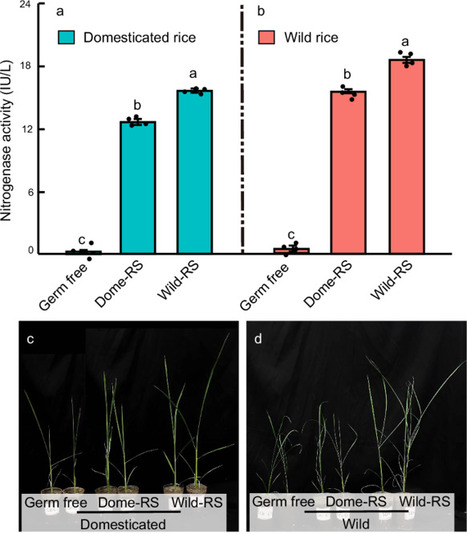
Crop domestication has revolutionized food production but increased agriculture’s reliance on fertilizers and pesticides. We investigate differences in the rhizosphere microbiome functions of wild and domesticated rice, focusing on nitrogen (N) cycling genes. Shotgun metagenomics and real-time PCR reveal a higher abundance of N-fixing genes in the wild rice rhizosphere microbiomes. Validation through transplanting rhizosphere microbiome suspensions shows the highest nitrogenase activity in soils with wild rice suspensions, regardless of planted rice type. Domesticated rice, however, exhibits an increased number of genes associated with nitrous oxide (N2O) production. Measurements of N2O emissions in soils with wild and domesticated rice are significantly higher in soil with domesticated rice compared to wild rice. Comparative root metabolomics between wild and domesticated rice further show that wild rice root exudates positively correlate with the frequency and abundance of microbial N-fixing genes, as indicated by metagenomic and qPCR, respectively. To confirm, we add wild and domesticated rice root metabolites to black soil, and qPCR shows that wild rice exudates maximize microbial N-fixing gene abundances and nitrogenase activity. Collectively, these findings suggest that rice domestication negatively impacts N-fixing bacteria and enriches bacteria that produce the greenhouse gas N2O, highlighting the environmental trade-offs associated with crop domestication.

|
Scooped by
Jean-Michel Ané
November 6, 9:44 AM
|
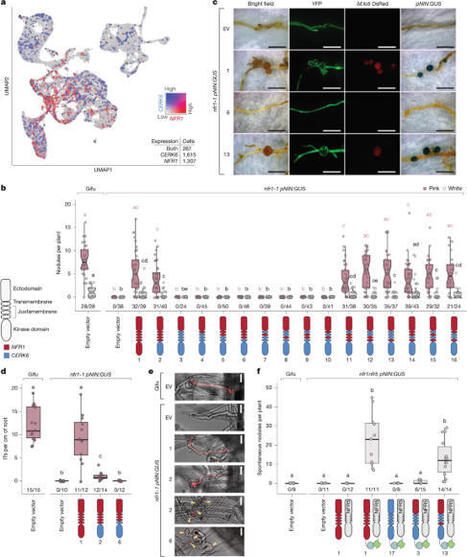
Receptor signalling determines cellular responses and is crucial for defining specific biological outcomes. In legume root cells, highly similar and structurally conserved chitin and Nod factor receptor kinases activate immune or symbiotic pathways, respectively, when chitinous ligands are perceived1. Here we show that specific amino acid residues in the intracellular part of the Nod factor receptor NFR1 control signalling specificity and enable the distinction of immune and symbiotic responses. Functional investigation of CERK6, NFR1 and receptor variants thereof revealed a conserved motif that we term Symbiosis Determinant 1 in the juxtamembrane region of the kinase domain, which is key for symbiotic signalling. We show that two residues in Symbiosis Determinant 1 are indispensable hallmarks of NFR1-type receptors and are sufficient to convert Lotus CERK6 and barley RLK4 kinase outputs to enable symbiotic signalling in Lotus japonicus.
|






 Your new post is loading...
Your new post is loading...