 Your new post is loading...
 Your new post is loading...

|
Scooped by
Jean-Michel Ané
February 10, 2:26 PM
|
Flavonoids, produced by the plant under nutrient stress, are required to initiate the legume-rhizobia symbiosis through the activation of rhizobial nod genes. Notwithstanding the central role of flavonoids in nodulation, their transcriptional regulation remains poorly understood. Here, we show that the nodulation signaling pathway 2 (NSP2) is required for transcriptional activation of flavonoid biosynthesis genes during nodulation in Medicago truncatula. Furthermore, MYB40, a legume-specific MYB transcription factor, is induced by rhizobia in the root epidermis. MYB40 directly binds to flavonoid biosynthetic gene promoters and is required for normal levels of nodulation. Biochemical and genetic evidence reveal that NSP2, not NSP1, interacts with MYB40 during rhizobial infection to strongly upregulate the symbiotic gene chalcone O-methyltransferase 1 in a manner dependent on MYB40 binding sites. Moreover, the overexpression of MYB40 and a microRNA-resistant NSP2 variant enhances nodulation under suboptimal rhizobial availability, suggesting this module fine-tunes symbiosis efficiency. Additionally, flavonoid regulation by NSP2 and MYB40 appears to facilitate arbuscular mycorrhizal colonization under nutrient starvation. Together, our findings establish an NSP2-MYB40 module that integrates symbiotic signaling with metabolic reprogramming, representing an evolutionary innovation for optimizing nitrogen acquisition in dynamic environments.

|
Scooped by
Jean-Michel Ané
February 6, 6:38 PM
|
Nitrogen is a major limiting nutrient for plant growth and a central driver of fertilizer use in agriculture. In many agricultural soils, nitrate is the main form of available nitrogen, but it is highly mobile and unevenly distributed, forcing plants to continuously adjust root growth to local supply. Legumes add an extra layer of complexity because they can also obtain ammonium through symbiotic nitrogen fixation in root nodules, which are carbon-expensive organs that must be regulated according to both external nitrate levels and internal nitrogen status. This review compares nitrogen signalling in the non-legume Arabidopsis thaliana and the model legume Medicago truncatula. Our aim is to understand how conserved pathways have been rewired to support these different nitrogen acquisition strategies. We focus on three core modules: CLE–CLV1/SUNN signalling, the NIN–NLP transcriptional module and the miR2111–TML pathway. In Arabidopsis, CLE–CLV1 acts mainly as a local low-nitrogen checkpoint that restricts lateral root emergence in nitrate-poor zones, while NLP7 integrates nitrate availability with root architectural responses. In Medicago, orthologous components have been recruited into the autoregulation of nodulation (AON) pathway, where nitrate-induced CLE peptides, NIN–NLP interactions and the systemic miR2111–TML module together integrate nitrate supply and rhizobial infection to coordinate nodulation with whole-plant nitrogen status and carbon cost. Within this network, the CLV1 ortholog SUNN acts as a central integration point. It responds to rhizobia-induced CLE peptides and to nitrogen-dependent systemic signals, including nitrate-induced CLE peptides, that influence lateral root growth in the absence of symbiosis. By contrasting these modules in Arabidopsis and Medicago, we show how legumes have layered symbiotic regulation onto ancestral nitrogen-signalling circuits that were first characterised in non-legume nitrate responses. This evolutionary perspective provides a mechanistic basis for future efforts to improve nitrogen use efficiency in crops by adjusting how CLE signalling, NIN/NLP activity and miR2111–TML-like modules link nitrogen status to root and nodule development.

|
Scooped by
Jean-Michel Ané
February 6, 6:22 PM
|
Nitrogen-fixing root nodule symbiosis (RNS) occurs in some eudicots, including legumes, and is regulated by the transcription factor NODULE INCEPTION (NIN), derived from the NIN-LIKE PROTEIN (NLP) family. However, how the NIN protein acquired RNS-specific functions remains unclear. We identify a previously undescribed motif in Lotus japonicus NIN, located downstream of the RWP-RK domain, which we term the FR. This motif broadens NIN’s DNA binding specificity by stabilizing the RWP-RK dimer interface. nin mutants lacking the FR motif show defective nodulation and impaired nitrogen fixation. Arabidopsis NLP2 carries a NIN-type FR and shares key features with NIN. Furthermore, the NIN-type FR had already emerged before the divergence of gymnosperm and angiosperm lineages, suggesting that a specific molecular feature of NIN involved in RNS regulation was inherited from ancestral NLPs prior to the emergence of RNS.

|
Scooped by
Jean-Michel Ané
February 4, 3:48 PM
|
•
A single origin of the RNS regulatory network, followed by lineage-specific refinements
•
Reconstructing the stepwise evolution of the RNS gene regulatory network
•
Paleohexaploidy at ∼110 mya provided founder genes crucial for later GRN-RNS assembly
•
Modules for cell wall remodeling and kinase signaling enable symbiosome formation

|
Scooped by
Jean-Michel Ané
February 4, 3:14 PM
|
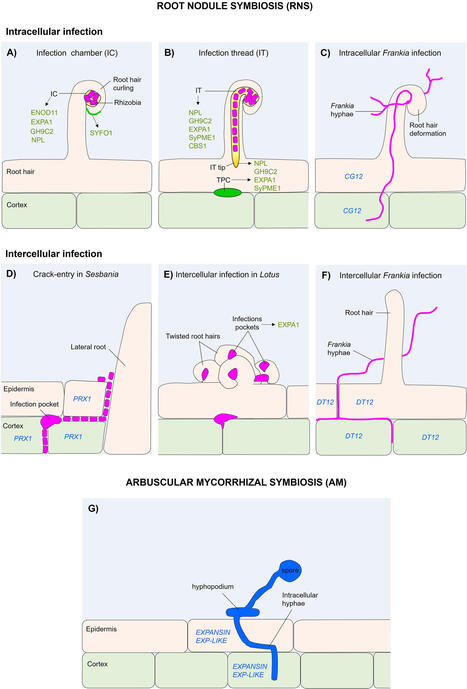
Plant roots are usually ground organs that perform essential roles, mostly associated with the anchoring of plants to the soil and absorption of nutrients and water. However, they are also exposed to a wide variety of microorganisms and may develop various symbiotic relationships, such as mutualism, which benefits both organisms. For instance, arbuscular mycorrhizal symbiosis is likely the oldest and most widespread mutualistic association, that occurs between plants and fungi. Another relevant example is the root nodule symbiosis, established between nitrogen-fixing bacteria and nodulating legumes, actinorhizal plants and Parasponia species. In both cases, microbial colonization of plant roots culminates in the formation of specialized symbiotic structures. In this regard, microbial infection is a critical step for the mutualistic relationship, where altering the cell wall biomechanics is necessary to facilitate microbial entry, which can be modulated by various cell wall protein families. This review examines the current knowledge on cell wall modifications occurring in plants roots during the symbiotic entry of microorganisms, focusing on the role of cell wall-remodeling proteins involved in these processes.

|
Scooped by
Jean-Michel Ané
January 31, 10:16 PM
|
Legume nodulation enables biological nitrogen fixation but is strongly repressed by nitrate. NIN-like proteins (NLPs) mediate this nitrate response, yet how their activity is regulated remains unclear. Here, we demonstrate that SUMOylation—a reversible posttranslational modification—is essential for the transcriptional activity and protein–protein interactions of MtNLP1 in Medicago truncatula, independently of its nitrate-induced nuclear localization. This modification is conserved in other NLPs, including Arabidopsis thaliana NLP7. Moreover, knockdown of SUMOylation-machinery components disrupts nodulation, suggesting that additional regulators in the symbiotic pathway also depend on SUMOylation. This work identifies SUMOylation as a conserved regulatory mechanism integrating nitrate signaling with root nodule symbiosis, with broad implications for improving plant nitrogen use efficiency.

|
Scooped by
Jean-Michel Ané
January 31, 9:04 PM
|
The cell surface localized lysin motif (LysM) receptor-like kinases/proteins (RLKs/RLPs) function as sensors for pathogenic and symbiotic microbes in land plants, perceiving chitin, lipo-chitooligosaccharides (LCOs), and peptidoglycan. LysM-RLKs/RLPs play a crucial role in activating various responses that lead to defense against pathogens or the establishment of symbiosis. While the functions of LysM-RLKs/RLPs were well-studied in land plants, their evolutionary origin and broader functional roles remain less explored. Streptophyte algae are widely recognized as the sister lineage of land plants. Land plants are believed to have emerged from a streptophyte algal ancestor. Plant–pathogen interactions are ancient and have played a pivotal role in shaping the evolution and complexity of the plant innate immune system. Genomic analyses revealed the presence of LysM-RLKs in two streptophyte algal species, Charophyceae Chara braunii and Zygnematophyceae Spirogyra pratensis. The functional roles of LysM-RLKs in both species were subsequently characterized. Through phylogenetic analysis combined with sequence alignment, I propose that LysM-RLKs originated from the last common ancestor of Charophyceae, Zygnematophyceae, and land plants. A conserved CXC motif within the LysM ectodomains (ECDs) was identified in streptophyte algal LysM-RLKs, like those present in land plants. Structural predictions using AlphaFold2 and three-dimensional modeling revealed that the overall architecture of LysM ECD is conserved, including a chitin-binding groove within the LysM2 domain. In vitro binding assays further demonstrated the chitin-binding capability of LysM ECDs from both streptophyte algae and bryophytes, suggesting an ancestral role of LysM ECDs in chitin recognition. Intriguingly, however, chitin treatment did not trigger downstream transcriptional responses in streptophyte algae, pointing to a functional divergence in LysM-RLKs during evolution. Furthermore, genetic and interaction studies demonstrated that heterodimerization and the formation of higher-order oligomeric complexes of LysM receptors are essential for proper function in bryophytes. In contrast to bryophytes, chitin treatment did not promote homodimerization of algal LysM receptors. Overall, this study new provides an insight into the evolutionary origin and functional diversification of LysM RLKs/RLPs in plants.

|
Scooped by
Jean-Michel Ané
January 31, 12:36 PM
|
Nitrogen isotope fractionation (ε15N) in sedimentary rocks has provided evidence for biological nitrogen fixation, and thus primary productivity, on the early Earth. However, the extent to which molecular evolution has influenced the isotopic signatures of nitrogenase, the enzyme that catalyzes the conversion of atmospheric nitrogen (N₂) to bioavailable ammonia, remains unresolved. Here, we reconstruct and experimentally characterize a library of synthetic ancestral nitrogenase genes, spanning over 2 billion years of evolutionary history. We assess the resulting ε¹⁵N values under controlled laboratory conditions. All engineered strains exhibit ε15N values within a narrow range comparable to that of modern microbes, suggesting that molybdenum (Mo)-dependent nitrogenase has been largely invariant throughout evolutionary time since the origins of this pathway. The results of this study support the early origin of molybdenum nitrogenase and the resilience of nitrogen-isotope biosignatures in ancient rocks, while also demonstrating their potential as powerful tools in the search for life beyond Earth.

|
Scooped by
Jean-Michel Ané
January 26, 7:04 PM
|
Arbuscular mycorrhizal fungi (AMF) establish a dynamic and asynchronous symbiosis with a wide range of land plants, involving distinct stages of root colonization and associated cellular responses that co-occur within the same root. Whilst decades of research have significantly advanced our understanding of the plant's symbiotic gene repertoire, this spatial and temporal complexity has hindered a detailed dissection of the molecular mechanisms underlying fungal accommodation. Here, we present the first single-nucleus RNA-sequencing (snRNA-seq) dataset of Solanum lycopersicum roots colonized by Rhizophagus irregularis. Unsupervised subclustering of an AM-specific cell population resolves AM-responsive root epidermal cells as well as a developmental gradient of cortical cells across distinct stages of arbuscule formation, unveiling stage-specific transcriptional signatures during AMF colonization. Moreover, using Motif-Informed Network Inference based on single-cell EXpression data (MINI-EX), we put forward candidate transcription factors orchestrating these stage-specific transcriptional programs. Together, our data support novel hypotheses on how diverse plant developmental and physiological processes – including localized cell cycle reactivation and the integration of multiple nutritional cues – are coordinated to facilitate the establishment of a functional symbiosis. As such, this high-resolution dataset serves as a valuable resource for candidate gene prioritization and future reverse genetic studies.

|
Scooped by
Jean-Michel Ané
January 26, 5:06 PM
|
Legumes are important sources of dietary protein and are key crops for sustainable agriculture because they fix atmospheric nitrogen via symbiotic interactions with rhizobia bacteria. However, legume plants are particularly sensitive to salt stress, with salinity negatively affecting development of the root nodule symbiosis. Genes that control salt-symbiosis crosstalk or trade-offs are largely unknown and poorly characterised. To assess the role of symbiosis signalling genes in salt stress, we analysed wildtype and symbiosis signalling mutants of Medicago truncatula grown in the presence of NaCl, sorbitol and/or rhizobia bacteria. We assessed root growth, plant biomass, nodule number and gene expression responses in plants exposed to stress. Our findings demonstrate that several symbiosis signalling genes play a previously undescribed role in regulating root responses to salt stress, including a calcium- and calcium/calmodulin-dependent protein kinase (CCaMK) and its interacting partner and downstream transcription factor, IPD3. Our results also show that the identified responses to salt stress are due to sodium toxicity rather than osmotic stress. We conclude that symbiosis signalling genes, including the CCaMK-IPD3 signalling module, may mediate signalling crosstalk between salt stress and symbiosis. These findings open new research avenues to explore how the environment regulates the legume-Rhizobium symbiosis.

|
Scooped by
Jean-Michel Ané
January 26, 4:52 PM
|

|
Scooped by
Jean-Michel Ané
January 19, 5:41 PM
|
Arbuscular mycorrhizal fungi (AMF) are widespread plant symbionts that enhance nutrient acquisition and influence ecosystem productivity. Previous chromosome-level assemblies of a model species revealed a two-compartment genome architecture (active A and repressed B chromatin compartments), yet its conservation across evolutionarily distant AMF lineages remains unresolved. Here, we present a chromosome-scale and 3D genome assembly of Gigaspora margarita isolate BEG34—the largest and most repeat-rich AMF genome to date—alongside that of its obligate endobacterium, Candidatus Glomerobacter gigasporarum (CaGg), using PacBio HiFi and Hi-C sequencing. The G. margarita genome comprises 43 chromosomes (792 Mb) organized into stable A/B compartments and Topologically Associating Domains structures, irrespective of the presence of endobacteria. We uncover 21 divergent rDNA operons distributed across six chromosomes and show that these physically interact, suggesting conserved nucleolar organization. We also reveal that the CaGg genome is tripartite and mobilome-rich, encoding prophages, an orphan CRISPR array, and complete pathways for many novel and essential cofactors, including heme, which may enhance host bioenergetics. We also find that the endobacterium’s presence regulates transposable elements in G. margarita. These findings reveal conserved principles of chromatin architecture in AMF symbionts and highlight the tight molecular interplay between fungal hosts and their endosymbionts, offering new insights into genome evolution and symbiotic adaptation.

|
Scooped by
Jean-Michel Ané
January 19, 5:34 PM
|
Nitrogen-fixing microbes are a primary contributor of this important nutrient to the global nitrogen cycle. Biological nitrogen fixation (BNF) through the enzyme nitrogenase requires extensive energy that in whole cells is generally studied during the oxidation of carbohydrates such as sugars. The nitrogen-fixing bacterium Azotobacter vinelandii is a model diazotroph for the study of aerobic BNF. Much is known about metabolism in A. vinelandii when cultured on a simple medium where energy is provided primarily in the form of sucrose or glucose. Outside of the laboratory, this soil bacterium grows on metabolites primarily derived from plant root exudates or from the degradation of dead plant matter. In this work, we expand on previous studies looking at genes that are essential to BNF in A. vinelandii when grown on sucrose medium using transposon sequencing (Tn-seq). We applied Tn-seq to determine the genes essential to growth when the medium was shifted to acetate, succinate or glycerol as the primary carbon and energy source to fuel both growth and BNF. A global overview of the genes of central metabolism and those directing substrates toward central metabolism, along with a selection of unexpected genes that were essential for specific growth substrates, is provided.
|

|
Scooped by
Jean-Michel Ané
February 10, 11:00 AM
|
Pesticides are widely distributed in soils1,2,3, yet their effects on soil biodiversity remain poorly understood4,5,6,7. Here we examined the effects of 63 pesticides on soil archaea, bacteria, fungi, protists, nematodes, arthropods and key functional gene groups across 373 sites spanning woodlands, grasslands and croplands in 26 European countries. Pesticide residues were detected in 70% of sites and emerged as the second strongest driver of soil biodiversity patterns after soil properties. Our analysis further revealed organism- and function-specific patterns, emphasizing complex and widespread non-target effects on soil biodiversity. Pesticides altered microbial functions, including phosphorus and nitrogen cycling, and suppressed beneficial taxa, including arbuscular mycorrhizal fungi and bacterivore nematodes. Our findings highlight the need to integrate functional and taxonomic characteristics into future risk assessment methodology to safeguard soil biodiversity, a cornerstone of ecosystem functioning.

|
Scooped by
Jean-Michel Ané
February 6, 6:36 PM
|
Arbuscular mycorrhizal (AM) fungi are known to enhance plant drought tolerance, but the physiological mechanism behind this benefit remains unclear. One explanation is that AM colonization improves root hydraulic conductance (Kr), thereby facilitating more efficient water uptake under soil drying, though this mechanism remains highly debated. Here, we measured Kr in tomato (Solanum lycopersicum L.) and pea (Pisum sativum L.) with and without AM using a noninvasive rehydration technique under soil drying, and this was complemented with the evaporative flux method under hydrated conditions. AM colonization was manipulated either through soil sterilization or by using nonmycorrhizal mutants, ensuring precise control of AM status. In both species, AM colonization had no positive impact on Kr under both well-hydrated and drought conditions. The finding suggests that the improved drought performance often observed in AM-colonized plants is not due to enhanced root water transport capacity. Instead, AM-induced benefits under drought may be mediated by other physiological adjustments.

|
Scooped by
Jean-Michel Ané
February 4, 3:52 PM
|
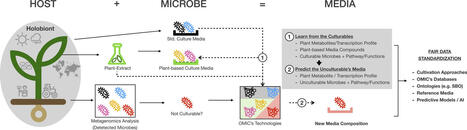
Background
The holobiont" refers to the plant and its associated microbiota that are pivotal to the plant's health, fitness, and survival. By in vitro culturing and functionally characterizing members of the plant microbiota, their specific roles in influencing plant responses to environmental changes can be determined and manipulated to foster sustainable agriculture and ecosystem management.
Aims
The review presents a comprehensive survey and current updates on culturomics of plant microbiota within the overall context of: a) the importance of understanding the plant holobiont composition and functioning; b) the necessity to in vitro track down and explore environmental microbiomes, entailing the plant microbiome with its myriad composition and spatio-temporal dynamics and mobility in various plant species, compartments and growth stages and c) the recent developments of the emerging in-situ similis cultivation strategies grounded on plant-based culture media.
Conclusions
The review highlights the urgent need to explore in vitro cultivation strategies built on compatible plant-based culture media, and the transformative role of omics technologies in refining these strategies. By bridging fundamental research and cultivation-based applications, such tools offer a gateway towards more sustainable and efficient in vitro cultivation systems, leading to a deeper understanding and potential manipulation of the plant holobiont.

|
Scooped by
Jean-Michel Ané
February 4, 3:37 PM
|
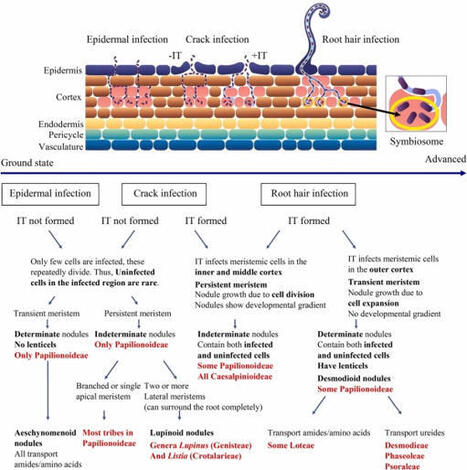
The Leguminosae (Fabaceae) is one of the largest and most evolutionary diverse plant families that comprises emblematic examples of symbiotic nitrogen fixation. This striking diversity is reflected across research areas ranging from taxonomy and phylogenomics to symbiosis, microbiology, biogeography, and functional traits. Insights into the genetic and molecular bases of root nodule symbiosis (RNS) are rapidly accumulating, enabling advances in agriculture and the engineering of biological nitrogen fixation (BNF). Nevertheless, substantial gaps remain, particularly regarding non-model species and the ecological and evolutionary drivers of RNS. This review summarises current knowledge on legume RNS and highlights that while the trait has likely contributed to legume diversification, the evolutionary trajectory of nodulation is complex, involving multiple gains, losses, and variations in symbiotic strategies across lineages. Nodule morphology and organogenesis, including determinate and indeterminate types, reveal structural and functional differences that may influence adaptability and BNF efficiency, although direct comparisons under varying environmental conditions remain limited. Ecological traits, such as drought tolerance, seed dormancy, and specialised pollination and defence mechanisms, interact with RNS to facilitate survival across diverse habitats. Case studies on Lebeckia ambigua and soybean wild relatives demonstrate how insights from non-model legumes can contribute to sustainable agriculture by improving stress resilience, expanding symbiotic partnerships, and broadening the genetic base for crop improvement. Future research should expand to non-model species and systematically assess nodulation, symbiotic efficiency, and environmental responsiveness to fully harness the potential of legumes for ecological and agricultural applications.

|
Scooped by
Jean-Michel Ané
February 2, 9:03 PM
|
Agriculture is under pressure to provide food for a growing population and the feedstock required to drive the bioeconomy. Methods to breed and genetically modify plants are inadequate to keep pace. When engineering crops, traits are painstakingly introduced into plants one-at-a-time, combine unpredictably, and are continuously expressed. Synthetic biology is changing these paradigms with new genome construction tools, computer aided design (CAD), and artificial intelligence (AI). “Smart plants” contain circuits that respond to environmental change, alter morphology, or respond to threats. Further, the plant and associated microbes (fungi, bacteria, archaea) are now being viewed by genetic engineers as a holistic system. Historically, plant health has been enhanced by many natural and laboratory-evolved soil microbes marketed to enhance growth or provide nutrients, or pest/stress resistance. Synthetic biology has expanded the number of species that can be engineered, increased the complexity of engineered functions, controlled environmental release, and can assemble stable consortia. New CAD tools will manage genetic engineering projects spanning multiple plant genomes (nucleus, chloroplast, mitochondrion) and the thousands of genomes of associated bacteria/fungi. This review covers advanced genetic engineering techniques to drive the next agricultural revolution, as well as push plant engineering into new realms for manufacturing, infrastructure, sensing, and remediation.

|
Scooped by
Jean-Michel Ané
January 31, 10:08 PM
|
Flavonoids are major plant secondary metabolites that mediate diverse plant–microbe interactions, including ectomycorrhizal (ECM) symbioses. However, their regulatory roles during ECM development remain poorly understood. Here, we investigated whether inoculation with Suillus bovinus alters flavonoid biosynthesis in Pinus yunnanensis roots and assessed how these flavonoids on fungal growth and gene expression. We applied exogenous flavonoids to S. bovinus mycelia to investigate fungal transcriptional and metabolic responses. Following inoculation, differentially expressed genes in P. yunnanensis roots were significantly enriched in the flavonoid biosynthesis pathway. Key enzyme-coding genes, including PAL, CHS, CHI, F3H, and FLS, were upregulated, and this was associated with increased flavonoid accumulation and enhanced antioxidant capacity. In S. bovinus, exogenous flavonoids promoted mycelial growth and induced metabolic adjustments related to carbohydrate and amino acid utilization. Several small secreted protein-related genes showed transcriptional responses to flavonoid exposure, indicating potential transcriptional modulation, although their specific roles in symbiosis remain unclear. These findings indicate that flavonoids may contribute to reciprocal interactions between host roots and ECM fungi and provide a molecular basis for further investigation.

|
Scooped by
Jean-Michel Ané
January 31, 9:03 PM
|
The Haber–Bosch process made synthetic ammonia abundant and helped feed a growing population, but it also created a sustainability debt: Substantial fossil-energy demand, significant CO2 emissions, and pervasive nitrogen losses that drive eutrophication and N2O emissions. Biological nitrogen fixation (BNF) is attractive because nitrogenase reduces N2 to NH3 at ambient conditions, yet the enzyme’s oxygen lability, complex metallocluster biosynthesis, and high energy demand complicate implementation beyond its native microbial contexts. This review synthesises three routes to reduce fertiliser dependence: engineered nitrogen-fixing biofertilisers that excrete ammonium; plant-centred strategies that extend symbiosis or express nitrogenase components in organelles; and purified nitrogenase or biohybrid systems powered by renewable electricity or light. We emphasise biohybrid devices because they decompose the challenge into modular interfaces (enzyme, power, microreactor environment) but require credible solutions for wiring, stability, continuous operation, and product capture. Across routes, protein engineering for stability, interface tolerance, and electron delivery is a shared enabling lever for system-level impact.

|
Scooped by
Jean-Michel Ané
January 26, 7:05 PM
|
The association of plants and microorganisms is a major determinant that influences the plant health, uptake of nutrients, and resilience to climate change. The technological advancements in the fields of genomics, transcriptomics, proteomics, and metabolomics have enabled understanding of these symbiotic interactions at cellular and molecular levels. The identification of molecular mechanisms that underlie the mutualistic association between plants and different kinds of beneficial microbes such as mycorrhizal fungi, rhizobia, endophytes, and plant growth-promoting rhizobacteria has revealed major signaling pathways such as the common symbiosis signaling pathway, hormone crosstalk, and microbe-associated molecular patterns. Recent studies have demonstrated that the Common Symbiosis Signaling Pathway (CSSP) is essential and conserved among diverse plant species, and assumes an important role in plant symbiotic interactions. Microbial consortia, notwithstanding their broad potential, are strongly dependent on the context and their results vary according to factors such as microbial competition, host genotype, and soil heterogeneity, which in turn explain the inconsistencies that have been observed in the field. The partnerships between plants and microbes could lead to exciting transformation for agriculture that’s both sustainable and resilient to climate challenges.

|
Scooped by
Jean-Michel Ané
January 26, 5:08 PM
|
The early nodulin-like (ENODL) subfamily, part of the phytocyanin, arabinogalactan protein, and nodulin-like families, is involved in plant growth and stress resistance. However, its role in symbiotic nodulation remains poorly understood. In barrel medic (Medicago truncatula), we found MtENODL29 was strongly activated at the late stages of nodule development, particularly in the infection zone of nodules. Both RNA interference (RNAi) and mutation of MtENODL29 caused a considerable reduction in nodule numbers, an increase in cysteine protease activity, a dramatic decrease in leghemoglobin content, and signs of premature senescence in nodule cells, suggesting that disruption of MtENODL29 accelerates nodule aging. Transcriptome analysis of 7-dpi (day post inoculation) inoculated roots and 28-dpi nodules in enodl29 mutants showed significant downregulation of symbiotic genes, accompanied by differential expression of genes associated with lipid metabolism and transport. MtENODL29 mutation also negatively impacted plant growth and development. MtENODL29 bound to MtnsLTP (non-specific lipid transfer protein), MtKCR (very-long-chain 3-oxoacyl-CoA reductase), and MtSec61γ (gamma subunit of the translocase complex Sec61) through its ALR (arabinogalactan protein-like region) domain. MtENODL29 co-localized with these proteins in the plasma membrane and endoplasmic reticulum. Notably, MtnsLTP showed high expression in the nodules, similar to MtENODL29, while MtKCR and MtSec61γ were also highly expressed in the leaves and stems. These results suggest that MtENODL29 participates in membrane lipid modification and transport by interacting with MtnsLTP, MtKCR, and MtSec61γ, facilitating the formation of symbiosome membranes as alfalfa rhizobium (Sinorhizobium meliloti) strain 1021 are released into nodule cells. Moreover, MtENODL29 influences plant growth, highlighting its role in coordinating plant development and symbiosis.

|
Scooped by
Jean-Michel Ané
January 26, 5:04 PM
|
In the current era of global climate change, agriculture is under extreme pressure due to variety of abiotic stresses (extreme temperature conditions, prolonged drought situations, unexpected flooding, soil salinization, luxurious use of agrochemicals, depleting micro flora, metal and metalloid toxicity and war or warlike situations) and biotic stress factors (pathogens, pests and weeds), all seriously challenging the global food security (Dervash et al. 2024; Lahlali et al. 2025; Jiang et al. 2025; Zhang et al. 2025; Manghwar and Zaman 2024; Lin et al. 2023). Abiotic and biotic stresses significantly disrupt the vital physiological processes like net photosynthetic rate, uptake of essential and beneficial nutrients, water uptake and overall health of the plants and lead to serious yield penalties (Ashraf et al. 2009, 2012; Hakeem et al. 2013; Hasanuzzaman et al. 2019; Ozturk and Gul 2020; Akhtar et al. 2024; Wang et al. 2025a, b).

|
Scooped by
Jean-Michel Ané
January 26, 4:46 PM
|
Plant-associated microbial communities consist of plant holobiont and play an essential role in plant growth and development, yet their collective functions are not fully understood. Theoretically, microbiota can act as integrated consortia, conferring emergent properties beyond those of single species. Here, we show that the tomato rhizosphere microbiome, when stimulated by a Flavobacterium dauae, enhances plant growth by activating the phytosterol biosynthesis pathway in both the microbiota and the plant host, a function unattainable by individual microbial species. A reconstructed synthetic community, based on meta-transcriptome of plant microbiota, recapitulated this microbiome-driven activity upon stimulation by F. dauae. This synthetic community also restored the growth response in diverse sterol-deficient plant mutants. The microbial consortium exhibits multispecies biofilm formation and functional specialization among its members, constituting a microbial coalition that promotes plant growth. This study provides direct experimental evidence that plant microbiota function as a coordinated unit and orchestrate host plant development. We highlight this to be a plant holobiont function based on microbial community coalition in host plant.

|
Scooped by
Jean-Michel Ané
January 19, 5:39 PM
|
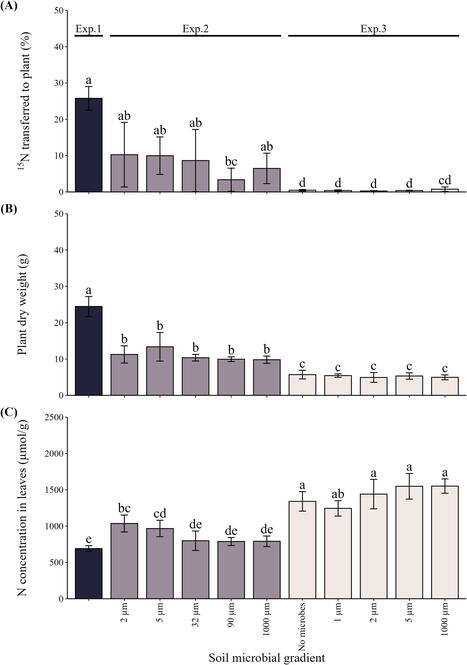
Bacground: Arbuscular mycorrhizal (AM) fungi enhance plant nutrient acquisition from soil; however, their ability to exploit organic nutrient forms in the absence of associated microbes capable of mineralization remains unclear.
Methods: To test if the AM fungi carry their beneficial bacterial partners into nutrient-rich zones, we conducted three controlled experiments manipulating the microbial inputs, diversity and composition in plant–AM fungus–soil systems, ranging from open pots to semi-sterile mesocosms. We manipulated soil microbial diversity by imposing a microbial diversity gradient (complex communities fractionated by size, resulting in fractions passing through 1 µm to 1000 µm sieves) and cultivated Andropogon gerardii in previously sterilized substrate together with a bacterial-free Rhizophagus irregularis. In each experiment, 15N‐labeled chitin or mineral nitrogen (N) compartments were installed in the root‐free zone of each mesocosm.
Results: With decreasing microbial inputs into the root-free zone, the N uptake from chitin to plants, facilitated by the AM fungal hyphae, decreased. Upon complete absence of microbes in the root-free zone, AM hyphal foraging preferences assessed by quantitative PCR indicated that exploration of the mineral N compartments was more effective than that of the chitin compartments. The AM fungal hyphae were ineffective in priming mineralization of organic N even if provided with complex soil microbiomes at a distance from the compartment.
Conclusions: In summary, chitin-enriched compartments become attractive for the AM fungi only when previously mineralized by competent microbes. Such microbes, however, were not effectively transported to spatially restricted organic resources in soil via AM hyphal highways in our experiments.
|






 Your new post is loading...
Your new post is loading...













Awesome paper. That's the surprise of the day!