 Your new post is loading...
 Your new post is loading...

|
Scooped by
NatProdChem
August 20, 2016 5:31 PM
|
Previously, we showed a few examples of preparing crude extracts from plants and other organisms. While sometimes (rarely) we get lucky and obtain an extract that is predominantly one compound, a more representative situation is that a mixture of compounds is obtained.

|
Scooped by
NatProdChem
August 10, 2016 1:27 AM
|
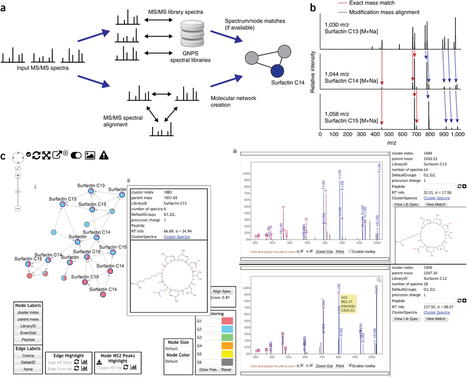
The potential of the diverse chemistries present in natural products (NP) for biotechnology and medicine remains untapped because NP databases are not searchable with raw data and the NP community has no way to share data other than in published papers. Although mass spectrometry (MS) techniques are well-suited to high-throughput characterization of NP, there is a pressing need for an infrastructure to enable sharing and curation of data. We present Global Natural Products Social Molecular Networking (GNPS; http://gnps.ucsd.edu), an open-access knowledge base for community-wide organization and sharing of raw, processed or identified tandem mass (MS/MS) spectrometry data. In GNPS, crowdsourced curation of freely available community-wide reference MS libraries will underpin improved annotations. Data-driven social-networking should facilitate identification of spectra and foster collaborations. We also introduce the concept of 'living data' through continuous reanalysis of deposited data.

|
Scooped by
NatProdChem
July 23, 2016 4:10 AM
|
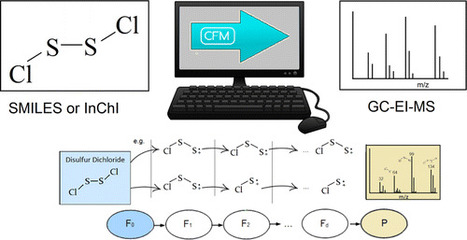
We describe a tool, competitive fragmentation modeling for electron ionization (CFM-EI) that, given a chemical structure (e.g., in SMILES or InChI format), computationally predicts an electron ionization mass spectrum (EI-MS) (i.e., the type of mass spectrum commonly generated by gas chromatography mass spectrometry). The predicted spectra produced by this tool can be used for putative compound identification, complementing measured spectra in reference databases by expanding the range of compounds able to be considered when availability of measured spectra is limited. The tool extends CFM-ESI, a recently developed method for computational prediction of electrospray tandem mass spectra (ESI-MS/MS), but unlike CFM-ESI, CFM-EI can handle odd-electron ions and isotopes and incorporates an artificial neural network. Tests on EI-MS data from the NIST database demonstrate that CFM-EI is able to model fragmentation likelihoods in low-resolution EI-MS data, producing predicted spectra whose dot product scores are significantly better than full enumeration “bar-code” spectra. CFM-EI also outperformed previously reported results for MetFrag, MOLGEN-MS, and Mass Frontier on one compound identification task. It also outperformed MetFrag in a range of other compound identification tasks involving a much larger data set, containing both derivatized and nonderivatized compounds. While replicate EI-MS measurements of chemical standards are still a more accurate point of comparison, CFM-EI’s predictions provide a much-needed alternative when no reference standard is available for measurement. CFM-EI is available at https://sourceforge.net/projects/cfm-id/ for download and http://cfmid.wishartlab.com as a web service. Department of Computing Science, University of Alberta, Edmonton T6G 2E8, Canada Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.6b01622 Publication Date (Web): July 06, 2016

|
Scooped by
NatProdChem
July 23, 2016 4:07 AM
|
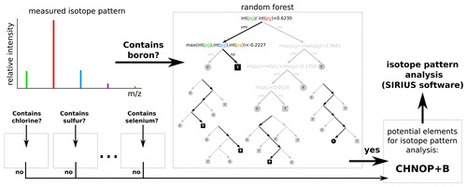
The determination of the molecular formula is one of the earliest and most important steps when investigating the chemical nature of an unknown compound. Common approaches use the isotopic pattern of a compound measured using mass spectrometry. Computational methods to determine the molecular formula from this isotopic pattern require a fixed set of elements. Considering all possible elements severely increases running times and more importantly the chance for false positive identifications as the number of candidate formulas for a given target mass rises significantly if the constituting elements are not prefiltered. This negative effect grows stronger for compounds of higher molecular mass as the effect of a single atom on the overall isotopic pattern grows smaller. On the other hand, hand-selected restrictions on this set of elements may prevent the identification of the correct molecular formula. Thus, it is a crucial step to determine the set of elements most likely comprising the compound prior to the assignment of an elemental formula to an exact mass. In this paper, we present a method to determine the presence of certain elements (sulfur, chlorine, bromine, boron, and selenium) in the compound from its (high mass accuracy) isotopic pattern. We limit ourselves to biomolecules, in the sense of products from nature or synthetic products with potential bioactivity. The classifiers developed here predict the presence of an element with a very high sensitivity and high specificity. We evaluate classifiers on three real-world data sets with 663 isotope patterns in total: 184 isotope patterns containing sulfur, 187 containing chlorine, 14 containing bromine, one containing boron, one containing selenium. In no case do we make a false negative prediction; for chlorine, bromine, boron, and selenium, we make ten false positive predictions in total. We also demonstrate the impact of our method on the identification of molecular formulas, in particular on the number of considered candidates and running time. The element prediction will be part of the next SIRIUS release, available from https://bio.informatik.uni-jena.de/software/sirius/. Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.6b01015 Publication Date (Web): July 11, 2016

|
Scooped by
NatProdChem
June 25, 2016 8:48 AM
|
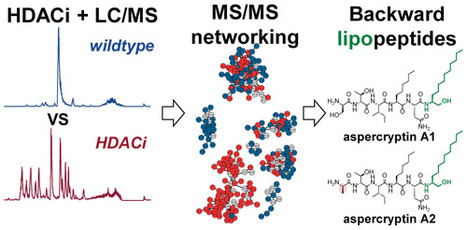
Unlocking the biochemical stores of fungi is key for developing future pharmaceuticals. Through reduced expression of a critical histone deacetylase in Aspergillus nidulans, increases of up to 100-fold were observed in the levels of 15 new aspercryptins, recently described lipopeptides with two noncanonical amino acids derived from octanoic and dodecanoic acids. In addition to two NMR-verified structures, MS/MS networking helped uncover an additional 13 aspercryptins. The aspercryptins break the conventional structural orientation of lipopeptides and appear “backward” when compared to known compounds of this class. We have also confirmed the 14-gene aspercryptin biosynthetic gene cluster, which encodes two fatty acid synthases and several enzymes to convert saturated octanoic and dodecanoic acid to α-amino acids. ACS Chem. Biol., Article ASAP DOI: 10.1021/acschembio.6b00398

|
Scooped by
NatProdChem
June 12, 2016 11:16 AM
|
Researchers now have the tools to unlock the potential of previously uncultivated microbes, prompting a return to drug discovery from natural products.

|
Scooped by
NatProdChem
June 2, 2016 3:55 AM
|
Covering: up to 2016" Dictazoles and sceptrins are singular metabolites of marine origin. The present dichotomic case study provides a comprehensive perspective on these cyclobutane-centered alkaloids and their respective families. Indeed, their upstream and downstream chemistry are both treated herein. Relevant isolation reports and bio-inspired total syntheses are used to decipher the currently admitted biosynthetic hypotheses as well as the emergence of diversity in the two series. This review proposes a transversal vision of the topic, where most aspects of natural product chemistry have a critical importance. Mehdi A. Beniddir,a Laurent Evanno,a Delphine Joseph,a Adam Skiredj*a and Erwan Poupon*a

|
Scooped by
NatProdChem
April 28, 2016 1:30 PM
|
Five new meroterpenoids, purpurogenolides A–E (1–5), and four known metabolites (6–9) were isolated from the solid substrate fermentation cultures of the fungus Penicillium purpurogenum MHz 111. The structures of the new meroterpenoids were elucidated by analysis of spectroscopic and spectrometric data (1D and 2D NMR, IR, and HRESIMS). The absolute configurations of 1 and 5 were determined by single-crystal X-ray crystallographic analysis, and those of 2–4 were elucidated on the basis of experimental and calculated electronic circular dichroism spectra. Compounds 2–4 and 6 showed inhibition of nitric oxide production in lipopolysaccharide-activated BV-2 microglial cells with IC50 values of 0.8–30.0 μM. Jing Sun†‡, Zhi-Xiang Zhu†, Yue-Lin Song†, Dan Dong§, Jiao Zheng†, Ting Liu§, Yun-Fang Zhao†, Daneel Ferreira⊥, Jordan K. Zjawiony⊥, Peng-Fei Tu*†, and Jun Li*† J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00160 Publication Date (Web): April 27, 2016

|
Scooped by
NatProdChem
April 15, 2016 1:48 AM
|
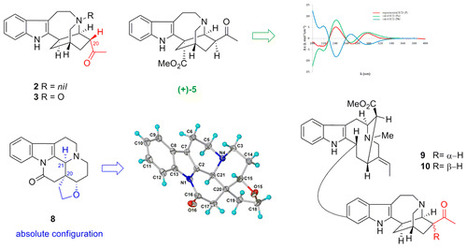
Click here to Ten new indole alkaloids (1–10) comprising five ibogan, two aspidosperman, one vincamine, and two bisindole alkaloids, in addition to 32 known alkaloids, were isolated from the stem-bark extract of a Malayan Tabernaemontana corymbosa. The structures of these alkaloids were determined based on analysis of the NMR and MS data and, in five instances (1, 3, 5, 6, 8), confirmed by X-ray diffraction analysis. Two of the iboga alkaloids, conodusines B (2) and C (3), and the iboga-containing bisindole tabernamidine B (10) are notable for the presence of an α-substituted acetyl group at C-20 of the iboga carbon skeleton. The iboga alkaloid (+)-conodusine E (5) had MS and NMR data that were identical to those of (−)-ervatamine I, recently isolated from Ervatamia hainanensis. Establishment of the absolute configuration of (+)-conodusine E (5) was based on analysis of the ECD data, correlation with (−)-heyneanine, and X-ray analysis, which showed that (+)-5 belongs to the same enantiomeric series as exemplified by (−)-coronaridine. The configuration at C-20′ of the previously reported Tabernaemontana bisindole alkaloid 19′-oxotabernamine (renamed tabernamidine B) required revision based on the present results. Several of the bisindoles showed pronounced in vitro growth inhibitory activity against drug-sensitive and vincristine-resistant KB cells. Department of Chemistry, Faculty of Science, University of Malaya, 50603 Kuala Lumpur, Malaysia J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00129

|
Scooped by
NatProdChem
March 30, 2016 6:03 AM
|
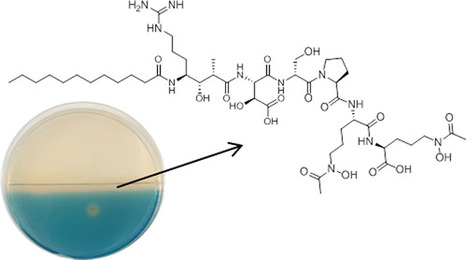
Photoreactive siderophores have a major impact on the growth of planktonic organisms. To date, these molecules have mainly been reported from marine bacteria, although evidence is now accumulating that some terrestrial bacteria also harbor the biosynthetic potential for their production. In this paper, we describe the genomics-driven discovery and characterization of variochelins, lipopeptide siderophores from the bacterium Variovorax boronicumulans, which thrives in soil and freshwater habitats. Variochelins are different from most other lipopeptide siderophores in that their biosynthesis involves a polyketide synthase. We demonstrate that the ferric iron complex of variochelin A possesses photoreactive properties and present the MS-derived structures of two degradation products that emerge upon light exposure. J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b00932

|
Scooped by
NatProdChem
March 21, 2016 2:09 AM
|
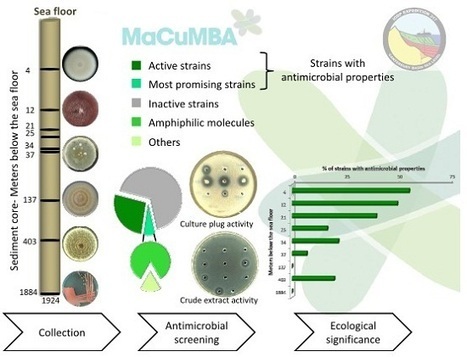
The evolving global threat of antimicrobial resistance requires a deep renewal of the antibiotic arsenal including the isolation and characterization of new drugs. Underexplored marine ecosystems may represent an untapped reservoir of novel bioactive molecules. Deep-sea fungi isolated from a record-depth sediment core of almost 2000 m below the seafloor were investigated for antimicrobial activities. This antimicrobial screening, using 16 microbial targets, revealed 33% of filamentous fungi synthesizing bioactive compounds with activities against pathogenic bacteria and fungi. Interestingly, occurrence of antimicrobial producing isolates was well correlated with the complexity of the habitat (in term of microbial richness), as higher antimicrobial activities were obtained at specific layers of the sediment core. It clearly highlights complex deep-sea habitats as chemical battlefields where synthesis of numerous bioactive compounds appears critical for microbial competition. The six most promising deep subseafloor fungal isolates were selected for the production and extraction of bioactive compounds. Depending on the fungal isolates, antimicrobial compounds were only biosynthesized in semi-liquid or solid-state conditions as no antimicrobial activities were ever detected using liquid fermentation. An exception was made for one fungal isolate, and the extraction procedure designed to extract amphipathic compounds was successful and highlighted the amphiphilic profile of the bioactive metabolites. Marion Navarri, Camille Jégou, Laurence Meslet-Cladière, Benjamin Brillet, Georges Barbier, Gaëtan Burgaudand Yannick Fleury *

|
Scooped by
NatProdChem
March 14, 2016 8:38 AM
|
Grisemycin (1), the first sulfur angucyclinone with an unusual ether-bridged system, was isolated from a marine-derived Streptomyces griseus strain M268. Its novel, here cage-like, structure was determined by spectroscopic analysis and single-crystal X-ray diffraction. Compound 1 exhibited modestly selective activity against the HL-60 cell line with an IC50 value of 31.54 μM. Futhermore, the absolute stereochemistry of kiamycin (2), an 1,12-epoxybenz[a]anthracene, previously obtained from the same strain, was established by X-ray diffraction analysis. Org. Lett., Article ASAP DOI: 10.1021/acs.orglett.6b00332 Publication Date (Web): March 9, 2016

|
Scooped by
NatProdChem
March 9, 2016 5:37 AM
|
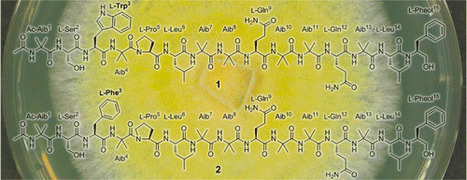
The Chilean Sepedonium aff. chalcipori strain KSH 883, isolated from the endemic Boletus loyo Philippi, was studied in a polythetic approach based on chemical, molecular, and biological data. A taxonomic study of the strain using molecular data of the ITS, EF1-α, and RPB2 barcoding genes confirmed the position of the isolated strain within the S. chalcipori clade, but also suggested the separation of this clade into three different species. Two new linear 15-residue peptaibols, named chilenopeptins A (1) and B (2), together with the known peptaibols tylopeptins A (3) and B (4) were isolated from the semisolid culture of strain KSH 883. The structures of 1 and 2 were elucidated on the basis of HRESIMSn experiments in conjunction with comprehensive 1D and 2D NMR analysis. Thus, the sequence of chilenopeptin A (1) was identified as Ac-Aib1-Ser2-Trp3-Aib4-Pro5-Leu6-Aib7-Aib8-Gln9-Aib10-Aib11-Gln12-Aib13-Leu14-Pheol15, while chilenopeptin B (2) differs from 1 by the replacement of Trp3 by Phe3. Additionally, the total synthesis of 1 and 2 was accomplished by a solid-phase approach, confirming the absolute configuration of all chiral amino acids as l. Both the chilenopeptins (1 and 2) and tylopeptins (3 and 4) were evaluated for their potential to inhibit the growth of phytopathogenic organisms. Alexander Otto†, Annegret Laub†, Lucile Wendt‡, Andrea Porzel†, Jürgen Schmidt†, Götz Palfner§, José Becerra§, Dirk Krüger⊥, Marc Stadler*‡, Ludger Wessjohann†∥, Bernhard Westermann*†∥, and Norbert Arnold*† J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.5b01018 Publication Date (Web): March 8, 2016
|

|
Scooped by
NatProdChem
August 18, 2016 9:08 AM
|
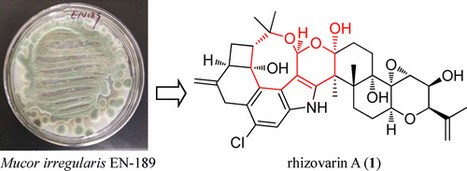
Genome mining of the fungus Mucor irregularis (formerly known as Rhizomucor variabilis) revealed the presence of various gene clusters for secondary metabolite biosynthesis, including several terpene-based clusters. Investigation into the chemical diversity of M. irregularis QEN-189, an endophytic fungus isolated from the fresh inner tissue of the marine mangrove plant Rhizophora stylosa, resulted in the discovery of 20 structurally diverse indole-diterpenes including six new compounds, namely, rhizovarins A–F (1–6). Among them, compounds 1–3 represent the most complex members of the reported indole-diterpenes. The presence of an unusual acetal linked to a hemiketal (1) or a ketal (2 and 3) in an unprecedented 4,6,6,8,5,6,6,6,6-fused indole-diterpene ring system makes them chemically unique. Their structures and absolute configurations were elucidated by spectroscopic analysis, modified Mosher’s method, and chemical calculations. Each of the isolated compounds was evaluated for antitumor activity against HL-60 and A-549 cell lines. J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00403

|
Scooped by
NatProdChem
July 23, 2016 4:13 AM
|
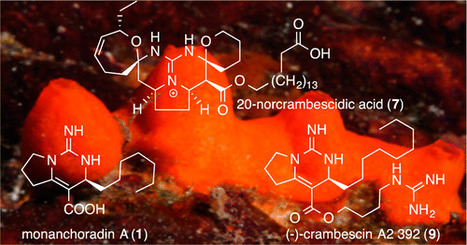
Four bicyclic and three pentacyclic guanidine alkaloids (1–7) were isolated from a French Polynesian Monanchora n. sp. sponge, along with the known alkaloids monalidine A (8), enantiomers 9–11 of known natural product crambescins, and the known crambescidins 12–15. Structures were assigned by spectroscopic data interpretation. The relative and absolute configurations of the alkaloids were established by analysis of 1H NMR and NOESY spectra and by circular dichroism analysis. The new norcrambescidic acid (7) corresponds to interesting biosynthetic variation within the pentacyclic core. All compounds exhibited antiproliferative and cytotoxic efficacy against KB, HCT116, HL60, MRC5, and B16F10 cancer cells, with IC50 values ranging from 4 nM to 10 μM. J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00168 Publication Date (Web): July 15, 2016

|
Scooped by
NatProdChem
July 23, 2016 4:08 AM
|
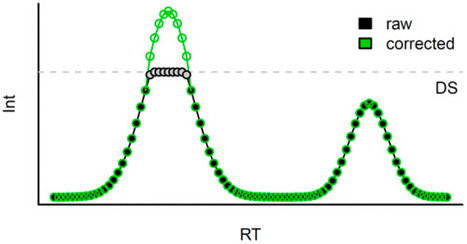
Metabolomics, the analysis of potentially all small molecules within a biological system, has become a valuable tool for biomarker identification and the elucidation of biological processes. While metabolites are often present in complex mixtures at extremely different concentrations, the dynamic range of available analytical methods to capture this variance is generally limited. Here, we show that gas chromatography coupled to atmospheric pressure chemical ionization mass spectrometry (GC-APCI-MS), a state of the art analytical technology applied in metabolomics analyses, shows an average linear range (LR) of 2.39 orders of magnitude for a set of 62 metabolites from a representative compound mixture. We further developed a computational tool to extend this dynamic range on average by more than 1 order of magnitude, demonstrated with a dilution series of the compound mixture, using robust and automatic reconstruction of intensity values exceeding the detection limit. The tool is freely available as an R package (CorrectOverloadedPeaks) from CRAN (https://cran.r-project.org/) and can be incorporated in a metabolomics data processing pipeline facilitating large screening assays. Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.6b02515

|
Scooped by
NatProdChem
July 11, 2016 3:50 AM
|
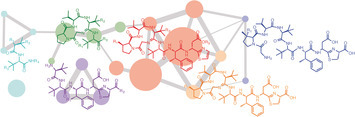
Bottromycin A2 is a structurally unique ribosomally synthesized and post-translationally modified peptide (RiPP) that possesses potent antibacterial activity towards multidrug-resistant bacteria. The structural novelty of bottromycin stems from its unprecedented macrocyclic amidine and rare β-methylated amino acid residues. The N-terminus of a precursor peptide (BtmD) is converted into bottromycin A2 by tailoring enzymes encoded in the btm gene cluster. However, little was known about key transformations in this pathway, including the unprecedented macrocyclization. To understand the pathway in detail, an untargeted metabolomic approach that harnesses mass spectral networking was used to assess the metabolomes of a series of pathway mutants. This analysis has yielded key information on the function of a variety of previously uncharacterized biosynthetic enzymes, including a YcaO domain protein and a partner protein that together catalyze the macrocyclization. William J. K. Crone, Natalia M. Vior, Javier Santos-Aberturas, Lukas G. Schmitz,
Finian J. Leeper, and Andrew W. Truman* Angewandte Chemistry International Edition: DOI: 10.1002/anie.201604304

|
Scooped by
NatProdChem
June 13, 2016 1:44 AM
|
« Humans can ingest gram amounts of plant secondary metabolites daily through diet. Many of these phytochemicals are bioactive beyond our current understanding because they act through weak negative biological feedback mechanisms, undetectable in vitro. Homeostatic-type assessments shed light on the evolutionary implications of the human diet from plants, giving rise to the metabolic plant feedback hypothesis. The hypothesis states that ancient diets rich in carbohydrates coincide with bulk dietary phytochemicals that act as nonspecific inhibitors of metabolic and inflammatory processes. Consequently, food-derived phytochemicals are likely to be equally effective as herbal medicines for these indications. In addition to the ubiquitous flavonoids, terpenoids, and fatty acids in the diet, the likely impact of chronic chlorophyll ingestion on human health is discussed, and data on its modulation of blood glucose levels are presented. A major deduction of this hypothesis is that starchy diets lacking plant secondary metabolites are associated with multimorbidity (lifestyle diseases) including obesity, type 2 diabetes, and cardiovascular disease. It is proposed that the intake of leafy vegetables, spices, and herbal remedies rich in phytochemicals matches the transition and genetic adaptation to early agriculture, playing a compensatory role in the mismatch of old genes and new diets. » Jürg Gertsch Planta Med
DOI: 10.1055/s-0042-108340

|
Scooped by
NatProdChem
June 7, 2016 12:26 PM
|
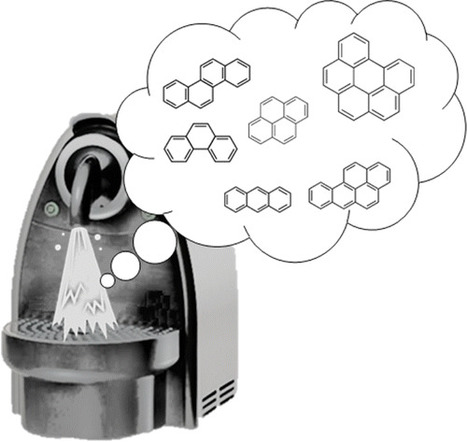
« A hard cap espresso machine has been used in combination with liquid chromatography with molecular fluorescence detection for the determination of polycyclic aromatic hydrocarbons (PAHs) from contaminated soils and sediments providing appropriate extraction efficiencies and quantitative results. Naphthalene, acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benz[a]anthracene, chrysene, benz[b]fluoranthene, benz[k]fluoranthene, benz[a]pyrene, dibenz[a,h]anthracene, benz[ghi]perylene, and indeno[1,2,3-cd]pyrene were used as target compounds. It should be mentioned that the pairs benz[a]anthracene–chrysene and dibenz[a,h]anthracene–benz[ghi]perylene peaks coelute under the employed chromatographic conditions; thus, those compounds were determined together. PAHs were extracted from 5.0 g of soil, previously homogenized, freeze-dried, and sieved to 250 μm, with 50 mL of 40% (v/v) acetonitrile in water at a temperature of 72 ± 3 °C. The proposed procedure is really fast, with an extraction time of 11 s, and it reduces the required amount of organic solvent to do the sample preparation. The obtained limit of detection for the evaluated PAHs was from 1 to 38 μg kg–1. Recoveries were calculated using clean soils spiked with 100, 500, 1000, and 2000 μg kg–1 PAHs with values ranging from 81 to 121% and good precision with relative standard deviation values lower than 30%. The method was validated using soil and sediment certified reference materials and also using real samples by comparison with ultrasound-assisted extraction, as reference methodology, obtaining statistically comparable results. Thus, the use of hard cap espresso machines in the analytical laboratories offers tremendous possibilities as low cost extraction units for the extraction of solid samples. » Hard Cap Espresso Machines in Analytical Chemistry: What Else?Department of Analytical Chemistry, University of Valencia, 50th Dr. Moliner Street, 46100 Burjassot, Spain Anal. Chem., Article ASAP DOI: 10.1021/acs.analchem.6b01400 Publication Date (Web): May 25, 2016 Copyright © 2016 American Chemical Society

|
Scooped by
NatProdChem
May 9, 2016 2:10 AM
|
Heteronuclear long-range NMR experiments are well established as essential NMR techniques for the structure elucidation of unknown natural products and small molecules. It is generally accepted that the absence of a given nJXH correlation in an HMBC or HSQMBC spectrum would automatically place the proton at least four bonds away from the carbon in question. This assumption can, however, be misleading in the case of a mismatch between the actual coupling constant and the delay used to optimize the experiment, which can lead to structural misassignments. Another scenario arises when an investigator, for whatever reason, needs to have access to very long-range correlations to confirm or refute a structure. In such cases, a conventional HMBC experiment will most likely fail to provide the requisite correlation, regardless of the delay optimization. Two recent methods for visualizing extremely weak or very long-range connectivities are the LR-HSQMBC and the HSQMBC-TOCSY experiments. Although they are intended to provide similar structural information, they utilize different transfer mechanisms, which differentiates the experiments, making each better suited for specific classes of compounds. In this report we have sought to examine the considerations implicit in choosing the best experiment to access weak or very long-range correlations for different types of molecules. J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00139 Publication Date (Web): May 3, 2016

|
Scooped by
NatProdChem
April 18, 2016 4:15 AM
|
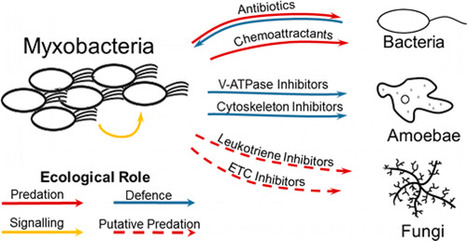
ClickThe study of natural products is entering a renaissance, driven by the discovery that the majority of bacterial secondary metabolites are not produced under standard laboratory conditions. Understanding the ecological role of natural products is key to efficiently directing our screening efforts, and to ensuring that each screen efficiently captures the full biosynthetic repertoire of the producing organisms. Myxobacteria represent one of the most common and diverse groups of bacteria, with roughly 2500 strains publically available. Fed largely through predation, the myxobacteria have developed a large repertoire of natural products that target other microorganisms, including bacteria and fungi. Many of these interactions can be observed in predation assays, providing direct evidence for environmental interactions. With a focus on Myxococcus xanthus, this review will highlight how recent advances in myxobacteria are revealing the chemical ecology of bacterial natural products. Department of Chemistry and Biochemistry, Faculty of Arts and Science, Concordia University, Montreal, Quebec, Canada H4B 1R6 ACS Chem. Biol., Article ASAP DOI: 10.1021/acschembio.6b00176

|
Scooped by
NatProdChem
April 13, 2016 4:09 PM
|
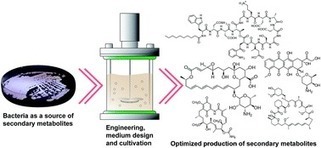
Metabolic engineering using systems biology tools is increasingly applied to overproduce secondary metabolites for their potential industrial production. In this Highlight, recent relevant metabolic engineering studies are analyzed with emphasis on host selection and engineering approaches for the optimal production of various prokaryotic secondary metabolites: native versus heterologous hosts (e.g., Escherichia coli) and rational versus random approaches. This comparative analysis is followed by discussions on systems biology tools deployed in optimizing the production of secondary metabolites. The potential contributions of additional systems biology tools are also discussed in the context of current challenges encountered during optimization of secondary metabolite production. Nat. Prod. Rep., 2016, Advance Article
DOI: 10.1039/C6NP00019C

|
Scooped by
NatProdChem
March 21, 2016 2:15 AM
|
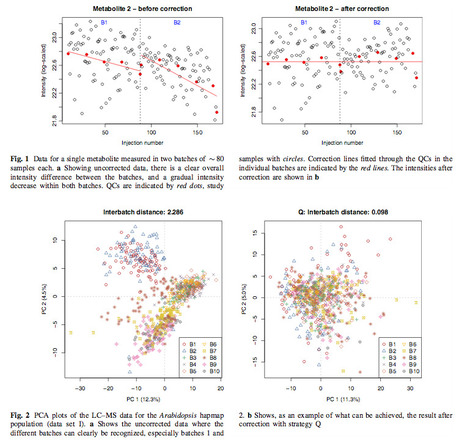
Introduction Batch effects in large untargeted metabolomics experiments are almost unavoidable, especially when sensitive detection techniques like mass spectrometry (MS) are employed. In order to obtain peak intensities that are comparable across all batches, corrections need to be performed. Since non-detects, i.e., signals with an intensity too low to be detected with certainty, are common in metabolomics studies, the batch correction methods need to take these into account. Objectives This paper aims to compare several batch correction methods, and investigates the effect of different strategies for handling non-detects. Methods Batch correction methods usually consist of regression models, possibly also accounting for trends within batches. To fit these models quality control samples (QCs), injected at regular intervals, can be used. Also study samples can be used, provided that the injection order is properly randomized. Normalization methods, not using information on batch labels or injection order, can correct for batch effects as well. Introducing two easy-to-use quality criteria, we assess the merits of these batch correction strategies using three large LC–MS and GC–MS data sets of samples from Arabidopsis thaliana. Results The three data sets have very different characteristics, leading to clearly distinct behaviour of the batch correction strategies studied. Explicit inclusion of information on batch and injection order in general leads to very good corrections; when enough QCs are available, also general normalization approaches perform well. Several approaches are shown to be able to handle non-detects—replacing them with very small numbers such as zero seems the worst of the approaches considered. Conclusion The use of quality control samples for batch correction leads to good results when enough QCs are available. If an experiment is properly set up, batch correction using the study samples usually leads to a similar high-quality correction, but has the advantage that more metabolites are corrected. The strategy for handling non-detects is important: choosing small values like zero can lead to suboptimal batch corrections. KeywordsBatch correction Untargeted metabolomics Non-detects Mass spectrometry Arabidopsis thaliana - Ron Wehrens
- , Jos. A. Hageman
- , Fred van Eeuwijk
- , Rik Kooke
- , Pádraic J. Flood
- , Erik Wijnker
- , Joost J. B. Keurentjes
- , Arjen Lommen
- , Henriëtte D. L. M. van Eekelen
- , Robert D. Hall
- , Roland Mumm
- , Ric C. H. de Vos
Metabolomics May 2016, 12:88

|
Scooped by
NatProdChem
March 17, 2016 6:57 AM
|
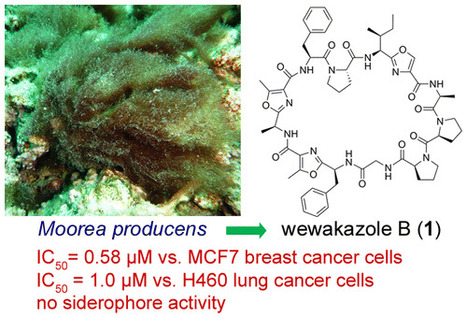
A mass spectrometry (MS)-guided isolation has led to the purification of a new cyanobactin, wewakazole B (1), along with the known compound curacin D from a Red Sea Moorea producens. The planar structure of 1 was elucidated using a combination of NMR and MS techniques. After ozonolysis and acid hydrolysis, the absolute configurations of the amino acid components of 1 were determined by chiral-phase LC-MS and HPLC analyses. Notably, compound 1 exhibited cytotoxic activity toward human MCF7 breast cancer cells (IC50 = 0.58 μM) and human H460 lung cancer cells (IC50 = 1.0 μM) and was also found to be inactive in a siderophore assay. J. Nat. Prod., Article ASAP DOI: 10.1021/acs.jnatprod.6b00051

|
Scooped by
NatProdChem
March 14, 2016 7:00 AM
|
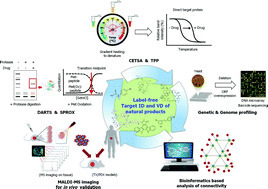
Identification of the target proteins of natural products is pivotal to understanding the mechanisms of action to develop natural products for use as molecular probes and potential therapeutic drugs. Affinity chromatography of immobilized natural products has been conventionally used to identify target proteins, and has yielded good results. However, this method has limitations, in that labeling or tagging for immobilization and affinity purification often result in reduced or altered activity of the natural product. New strategies have recently been developed and applied to identify the target proteins of natural products and synthetic small molecules without chemical modification of the natural product. These direct and indirect methods for target identification of label-free natural products include drug affinity responsive target stability (DARTS), stability of proteins from rates of oxidation (SPROX), cellular thermal shift assay (CETSA), thermal proteome profiling (TPP), and bioinformatics-based analysis of connectivity. This review focuses on and reports case studies of the latest advances in target protein identification methods for label-free natural products. The integration of newly developed technologies will provide new insights and highlight the value of natural products for use as biological probes and new drug candidates Nat. Prod. Rep., 2016, Advance Article DOI: 10.1039/C5NP00107B
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...
















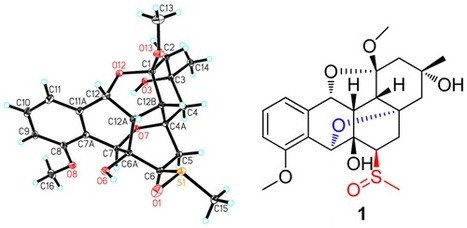







Excellent website MOC (Master Organic Chemistry) sums up some of Natural Products isolations techniques ... perfect for students ! Check out the crystal glamour shots !