Your new post is loading...
 Your new post is loading...

|
Rescooped by
Kamoun Lab @ TSL
from Plant Cell Biology
February 21, 2012 2:33 PM
|
I am very happy that Petra agreed to answer my questionnaire and tell us about her current research. Petra now works at the James Hutton Institute in Invergowrie, Dundee and is studying the interaction between plants and plant pathogens.
Via Anne Osterrieder

|
Scooped by
Kamoun Lab @ TSL
February 18, 2012 4:46 AM
|
Plants and microbes interact in a complex relationship that can have both harmful and beneficial impacts on both plant and microbial communities. Effectors, secreted microbial molecules that alter plant processes and facilitate colonization, are central to understanding the complicated interplay between plants and microbes. Effectors in Plant-Microbe Interactions unlocks the molecular basis of this important class of microbial molecules and describes their diverse and complex interactions with host plants.
Effectors in Plant Microbe Interactions is divided into five sections that take stock of the current knowledge on effectors of plant-associated organisms. Coverage ranges from the impact of bacterial, fungal and oomycete effectors on plant immunity and high-throughput genomic analysis of effectors to the function and trafficking of these microbial molecules. The final section looks at effectors secreted by other eukaryotic microbes that are the focus of current and future research efforts.
Written by leading international experts in plant-microbe interactions, Effectors in Plant Microbe Interactions, will be an essential volume for plant biologists, microbiologists, pathologists, and geneticists.

|
Scooped by
Kamoun Lab @ TSL
February 14, 2012 4:52 PM
|
Molecular Plant-Microbe Interactions invites original research manuscripts on the molecular biology and molecular genetics/genomics of parasitic interactions of nematodes and insects with plants for a special 2012 issue. This special issue will bring added attention to critically important research that has shown significant progress in recent years. Article Submission Deadline: May 21, 2012.

|
Scooped by
Kamoun Lab @ TSL
February 11, 2012 9:57 AM
|
The innate immune system is an ancient and broad-spectrum defense system found in all eukaryotes. The detection of microbial elicitors results in the up-regulation of defense-related genes and the elicitation of inflammatory and apoptotic responses. These innate immune responses are the front-line barrier against disease because they collectively suppress the growth of the vast majority of invading microbes. Despite their critical role, we know remarkably little about the diversity of immune elicitors. To address this paucity, we reasoned that hosts are more likely to evolve recognition to “core” pathogen proteins under strong negative selection for the maintenance of essential cellular functions, whereas repeated exposure to host–defense responses will impose strong positive selective pressure for elicitor diversification to avoid host recognition. Therefore, we hypothesized that novel bacterial elicitors can be identified through these opposing forces of natural selection. We tested this hypothesis by examining the genomes of six bacterial phytopathogens and identifying 56 candidate elicitors that have an excess of positively selected residues in a background of strong negative selection. We show that these positively selected residues are atypically clustered, similar to patterns seen in the few well-characterized elicitors. We then validated selected candidate elicitors by showing that they induce Arabidopsis thaliana innate immunity in functional (virulence suppression) and cellular (callose deposition) assays. These finding provide targets for the study of host–pathogen interactions and applied research into alternative antimicrobial treatments.

|
Scooped by
Kamoun Lab @ TSL
February 9, 2012 6:17 AM
|
Understanding plant–virus coevolution requires wild systems in which there is no human manipulation of either host or virus. To develop such a system, we analysed virus infection in six wild populations of Arabidopsis thaliana in Central Spain. The incidence of five virus species with different life-styles was monitored during four years, and this was analysed in relation to the demography of the host populations. Total virus incidence reached 70 per cent, which suggests a role of virus infection in the population structure and dynamics of the host, under the assumption of a host fitness cost caused by the infection. Maximum incidence occurred at early growth stages, and co-infection with different viruses was frequent, two factors often resulting in increased virulence. Experimental infections under controlled conditions with two isolates of the most prevalent viruses, cauliflower mosaic virus and cucumber mosaic virus, showed that there is genetic variation for virus accumulation, although this depended on the interaction between host and virus genotypes. Comparison of QST-based genetic differentiations between both host populations with FST genetic differentiation based on putatively neutral markers suggests different selection dynamics for resistance against different virus species or genotypes. Together, these results are compatible with a hypothesis of plant–virus coevolution.

|
Scooped by
Kamoun Lab @ TSL
February 8, 2012 6:28 AM
|
Knowledge about the distribution of mutational fitness effects (DMFE) is essential for many evolutionary models. In recent years, the properties of the DMFE have been carefully described for some microorganisms. In most cases, however, this information has been obtained only for a single environment, and very few studies have explored the effect that environmental variation may have on the DMFE. Environmental effects are particularly relevant for the evolution of multi-host parasites and thus for the emergence of new pathogens. Here we characterize the DMFE for a collection of twenty single-nucleotide substitution mutants of Tobacco etch potyvirus (TEV) across a set of eight host environments. Five of these host species were naturally infected by TEV, all belonging to family Solanaceae, whereas the other three were partially susceptible hosts belonging to three other plant families. First, we found a significant virus genotype-by-host species interaction, which was sustained by differences in genetic variance for fitness and the pleiotropic effect of mutations among hosts. Second, we found that the DMFEs were markedly different between Solanaceae and non-Solanaceae hosts. Exposure of TEV genotypes to non-Solanaceae hosts led to a large reduction of mean viral fitness, while the variance remained constant and skewness increased towards the right tail. Within the Solanaceae hosts, the distribution contained an excess of deleterious mutations, whereas for the non-Solanaceae the fraction of beneficial mutations was significantly larger. All together, this result suggests that TEV may easily broaden its host range and improve fitness in new hosts, and that knowledge about the DMFE in the natural host does not allow for making predictions about its properties in an alternative host.

|
Scooped by
Kamoun Lab @ TSL
February 6, 2012 4:56 PM
|
BTI scientists have released a draft sequence of the Nicotiana benthamiana genome which is accessible through the SGN BLAST tool and can be downloaded by ftp (see: http://solgenomics.net/). N. benthamiana is a widely used model for plant-microbe biology and other research applications. It is particularly useful because it is related to tomato and potato and is amenable to virus-induced gene silencing (VIGS) which facilitates the efficient functional study of plant genes.

|
Scooped by
Kamoun Lab @ TSL
February 5, 2012 7:48 AM
|
The last 20 years have provided a sophisticated understanding of how plants recognize relatively conserved microbial patterns to activate defence. This workshop will cover broad aspects of the plant-microbe interaction and train methods to study and visualise intracellular interactions during pathogenesis and defence.

|
Scooped by
Kamoun Lab @ TSL
February 2, 2012 12:15 PM
|
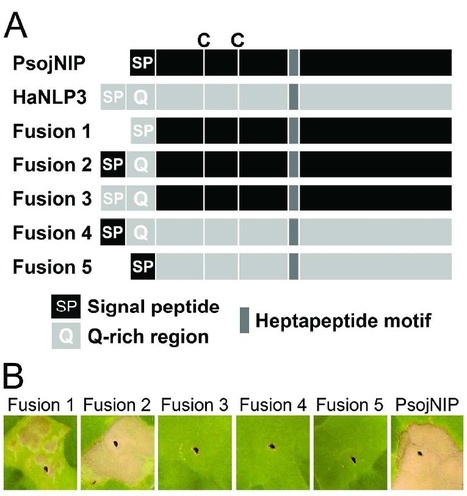
The genome of the downy mildew pathogen Hyaloperonospora arabidopsidis (Hpa) encodes Necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs). Although NLPs are widely distributed in eukaryotic and prokaryotic plant pathogens, it was surprising to find these proteins in the obligate biotrophic oomycete Hpa. We therefore analyzed the Hpa NLP (HaNLP) gene family and identified 12 HaNLPs and 15 pseudogenes. Most of the 27 genes form an Hpa-specific cluster when compared to other oomycete NLP genes, suggesting this class of effectors has recently expanded in Hpa. HaNLP transcripts were mainly detected during early infection stages. Agrobacterium tumefaciens-mediated transient expression and infiltration of recombinant NLPs into tobacco and Arabidopsis leaves revealed that all HaNLPs tested are non-cytotoxic proteins. Even HaNLP3 does not induce necrosis although it is the one most similar to necrosis-inducing NLPs of other oomycetes, and contains all amino acids that are critical for necrosis-inducing activity. Chimeras constructed between HaNLP3 and the necrosis-inducing PsojNIP protein demonstrated that most of the HaNLP3 protein is functionally equivalent to PsojNIP, except for an exposed domain that prevents necrosis-induction. The early expression and species-specific expansion of the HaNLPs is suggestive of an alternative function of non-cytolytic NLPs during biotrophic infection of plants.

|
Scooped by
Kamoun Lab @ TSL
February 1, 2012 12:12 PM
|
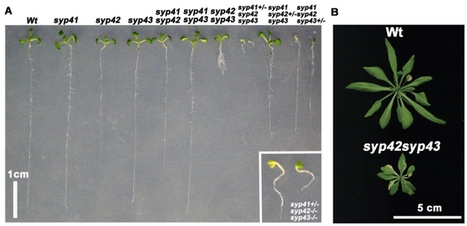
In all eukaryotic cells, a membrane-trafficking system connects the post-Golgi organelles, such as the trans-Golgi network (TGN), endosomes, vacuoles, and the plasma membrane. This complex network plays critical roles in several higher-order functions in multicellular organisms. The TGN, one of the important organelles for protein transport in the post-Golgi network, functions as a sorting station, where cargo proteins are directed to the appropriate post-Golgi compartments. Unlike its roles in animal and yeast cells, the TGN has also been reported to function like early endosomal compartments in plant cells. However, the physiological roles of the TGN functions in plants are not understood. Here, we report a study of the SYP4 group (SYP41, SYP42, and SYP43), which represents the plant orthologs of the Tlg2/syntaxin16 Qa-SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptor) that localizes on the TGN in yeast and animal cells. The SYP4 group regulates the secretory and vacuolar transport pathways in the post-Golgi network and maintains the morphology of the Golgi apparatus and TGN. Consistent with a secretory role, SYP4 proteins are required for extracellular resistance responses to a fungal pathogen. We also reveal a plant cell-specific higher-order role of the SYP4 group in the protection of chloroplasts from salicylic acid-dependent biotic stress.

|
Scooped by
Kamoun Lab @ TSL
January 25, 2012 9:50 AM
|
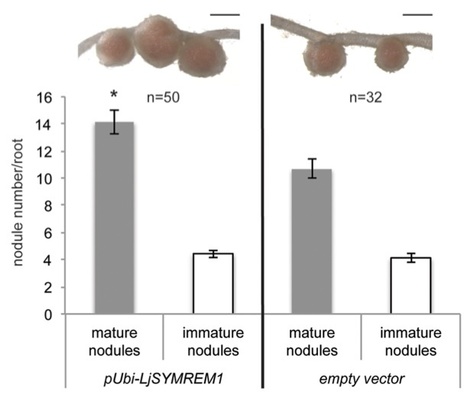
In legumes rhizobial infection during root nodule symbiosis (RNS) is controlled by a conserved set of receptor proteins and downstream components. MtSYMREM1, a protein of the Remorin family in Medicago truncatula, was shown to interact with at least three receptor-like kinases (RLKs) that are essential for RNS. Remorins are comprised of a conserved C-terminal domain and a variable N-terminal region that defines the six different Remorin groups. While both N- and C-terminal regions of Remorins belonging to the same phylogenetic group are similar to each other throughout the plant kingdom, the N-terminal domains of legume-specific group 2 Remorins show exceptional high degrees of sequence divergence suggesting evolutionary specialization of this protein within this clade. We therefore identified and characterized the MtSYMREM1 ortholog from Lotus japonicus (LjSYMREM1), a model legume that forms determinate root nodules. Here, we resolved its spatio-temporal regulation and showed that over-expression of LjSYMREM1 increases nodulation on transgenic roots. Using a structure-function approach we show that protein interactions including Remorin oligomerization are mainly mediated and stabilized by the Remorin C-terminal region with its coiled-coil domain while the RLK kinase domains transiently interact in vivo and phosphorylate a residue in the N-terminal region of the LjSYMREM1 protein in vitro. These data provide novel insights into the mechanism of this putative molecular scaffold protein and underline its importance during rhizobial infection.

|
Scooped by
Kamoun Lab @ TSL
January 16, 2012 6:18 PM
|
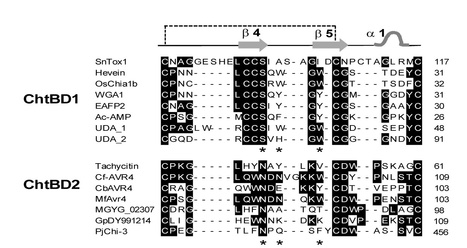
The wheat pathogen Stagonospora nodorum produces multiple necrotrophic effectors (also called host-selective toxins) that promote disease by interacting with corresponding host sensitivity gene products. SnTox1 was the first necrotrophic effector identified in S. nodorum, and was shown to induce necrosis on wheat lines carrying Snn1. Here, we report the molecular cloning and validation of SnTox1 as well as the preliminary characterization of the mechanism underlying the SnTox1-Snn1 interaction which leads to susceptibility. SnTox1 was identified using bioinformatics tools and verified by heterologous expression in Pichia pastoris. SnTox1 encodes a 117 amino acid protein with the first 17 amino acids predicted as a signal peptide, and strikingly, the mature protein contains 16 cysteine residues, a common feature for some avirulence effectors. The transformation of SnTox1 into an avirulent S. nodorum isolate was sufficient to make the strain pathogenic. Additionally, the deletion of SnTox1 in virulent isolates rendered the SnTox1 mutated strains avirulent on the Snn1 differential wheat line. SnTox1 was present in 85% of a global collection of S. nodorum isolates. We identified a total of 11 protein isoforms and found evidence for strong diversifying selection operating on SnTox1. The SnTox1-Snn1 interaction results in an oxidative burst, DNA laddering, and pathogenesis related (PR) gene expression, all hallmarks of a defense response. In the absence of light, the development of SnTox1-induced necrosis and disease symptoms were completely blocked. By comparing the infection processes of a GFP-tagged avirulent isolate and the same isolate transformed with SnTox1, we conclude that SnTox1 may play a critical role during fungal penetration. This research further demonstrates that necrotrophic fungal pathogens utilize small effector proteins to exploit plant resistance pathways for their colonization, which provides important insights into the molecular basis of the wheat-S. nodorum interaction, an emerging model for necrotrophic pathosystems.

|
Scooped by
Kamoun Lab @ TSL
January 10, 2012 10:24 AM
|
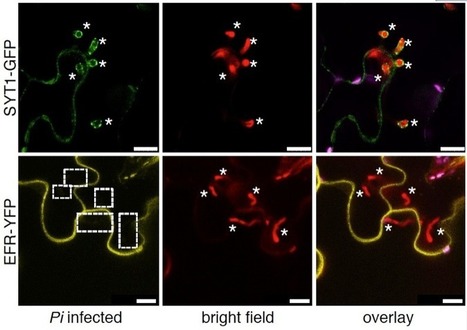
Adapted filamentous pathogens such as the oomycetes Hyaloperonospora arabidopsidis (Hpa) and Phytophthora infestans (Pi) project specialized hyphae, the haustoria, inside living host cells for the suppression of host defense and acquisition of nutrients. Accommodation of haustoria requires reorganization of the host cell and the biogenesis of a novel host cell membrane, the extrahaustorial membrane (EHM), which envelops the haustorium separating the host cell from the pathogen. Here, we applied live-cell imaging of fluorescent-tagged proteins labeling a variety of membrane compartments and investigated the subcellular changes associated with accommodating oomycete haustoria in Arabidopsis and N. benthamiana. Plasma membrane-resident proteins differentially localized to the EHM. Likewise, secretory vesicles and endosomal compartments surrounded Hpa and Pi haustoria revealing differences between these two oomycetes, and suggesting a role for vesicle trafficking pathways for the pathogen-controlled biogenesis of the EHM. The latter is supported by enhanced susceptibility of mutants in endosome-mediated trafficking regulators. These observations point at host subcellular defenses and specialization of the EHM in a pathogen-specific manner. Defense-associated haustorial encasements, a double-layered membrane that grows around mature haustoria, were frequently observed in Hpa interactions. Intriguingly, all tested plant proteins accumulated at Hpa haustorial encasements suggesting the general recruitment of default vesicle trafficking pathways to defend pathogen access. Altogether, our results show common requirements of subcellular changes associated with oomycete biotrophy, and highlight differences between two oomycete pathogens in reprogramming host cell vesicle trafficking for haustoria accommodation. This provides a framework for further dissection of the pathogen-triggered reprogramming of host subcellular changes.
|

|
Scooped by
Kamoun Lab @ TSL
February 19, 2012 2:24 PM
|
Hyaloperonospora arabidopsidis (Hpa) is an obligate biotroph oomycete pathogen of the model plant Arabidopsis thaliana and contains a large set of effector proteins that are translocated to the host to exert virulence functions or trigger immune responses. These effectors are characterized by conserved amino-terminal translocation sequences and highly divergent carboxyl-terminal functional domains. The availability of the Hpa genome sequence allowed the computational prediction of effectors and the development of effector delivery systems enabled validation of the predicted effectors in Arabidopsis. In this study, we identified a novel effector ATR39-1 by computational methods, which was found to trigger a resistance response in the Arabidopsis ecotype Weiningen (Wei-0). The allelic variant of this effector, ATR39-2, is not recognized, and two amino acid residues were identified and shown to be critical for this loss of recognition. The resistance protein responsible for recognition of the ATR39-1 effector in Arabidopsis is RPP39 and was identified by map-based cloning. RPP39 is a member of the CC-NBS-LRR family of resistance proteins and requires the signaling gene NDR1 for full activity. Recognition of ATR39-1 in Wei-0 does not inhibit growth of Hpa strains expressing the effector, suggesting complex mechanisms of pathogen evasion of recognition, and is similar to what has been shown in several other cases of plant-oomycete interactions. Identification of this resistance gene/effector pair adds to our knowledge of plant resistance mechanisms and provides the basis for further functional analyses.

|
Scooped by
Kamoun Lab @ TSL
February 17, 2012 3:31 AM
|
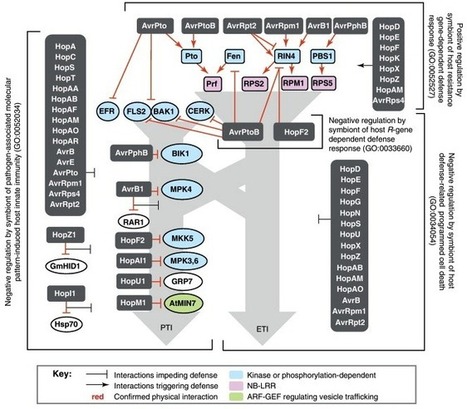
Many plant pathogens subvert host immunity by injecting compositionally diverse but functionally similar repertoires of cytoplasmic effector proteins. The bacterial pathogen Pseudomonas syringae is a model for exploring the functional structure of such repertoires. The pangenome of P. syringae encodes 57 families of effectors injected by the type III secretion system. Distribution of effector genes among phylogenetically diverse strains reveals a small set of core effectors targeting antimicrobial vesicle trafficking and a much larger set of variable effectors targeting kinase-based recognition processes. Complete disassembly of the 28-effector repertoire of a model strain and reassembly of a minimal functional repertoire reveals the importance of simultaneously attacking both processes. These observations, coupled with growing knowledge of effector targets in plants, support a model for coevolving molecular dialogs between effector repertoires and plant immune systems that emphasizes mutually-driven expansion of the components governing recognition.

|
Scooped by
Kamoun Lab @ TSL
February 13, 2012 5:01 PM
|
The orange and grapefruit harvests have begun again in the Rio Grande Valley, but the discovery of an outbreak of citrus greening disease there last month has raised concern. [Mamoudou Setamou, a citrus entomologist, shows greening's effects.]

|
Scooped by
Kamoun Lab @ TSL
February 9, 2012 11:08 AM
|
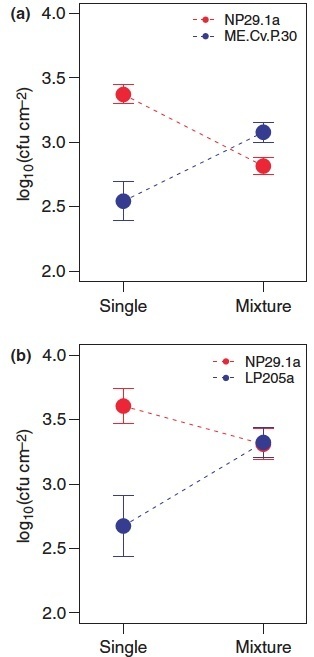
The evolutionary dynamics of pathogens are critically important for disease outcomes, prevalence and emergence. In this study we investigate ecological conditions that may promote the long-term maintenance of virulence polymorphisms in pathogen populations. Recent theory predicts that evolution towards increased virulence can be reversed if less-aggressive social ‘cheats’ exploit more aggressive ‘cooperator’ pathogens. However, there is no evidence that social exploitation operates within natural pathogen populations. We show that for the bacterium Pseudomonas syringae, major polymorphisms for pathogenicity are maintained at unexpectedly high frequencies in populations infecting the host Arabidopsis thaliana. Experiments reveal that less-aggressive strains substantially increase their growth potential in mixed infections and have a fitness advantage in non-host environments. These results suggest that niche differentiation can contribute to the maintenance of virulence polymorphisms, and that both within-host and between-host growth rates modulate cheating and cooperation in P. syringae populations.

|
Scooped by
Kamoun Lab @ TSL
February 9, 2012 4:40 AM
|
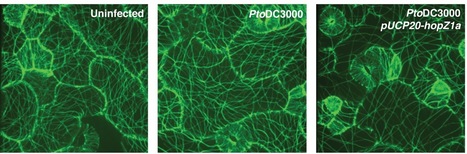
The eukaryotic cytoskeleton is essential for structural support and intracellular transport, and is therefore a common target of animal pathogens. However, no phytopathogenic effector has yet been demonstrated to specifically target the plant cytoskeleton. Here we show that the Pseudomonas syringae type III secreted effector HopZ1a interacts with tubulin and polymerized microtubules. We demonstrate that HopZ1a is an acetyltransferase activated by the eukaryotic co-factor phytic acid. Activated HopZ1a acetylates itself and tubulin. The conserved autoacetylation site of the YopJ / HopZ superfamily, K289, plays a critical role in both the avirulence and virulence function of HopZ1a. Furthermore, HopZ1a requires its acetyltransferase activity to cause a dramatic decrease in Arabidopsis thaliana microtubule networks, disrupt the plant secretory pathway and suppress cell wall-mediated defense. Together, this study supports the hypothesis that HopZ1a promotes virulence through cytoskeletal and secretory disruption.

|
Scooped by
Kamoun Lab @ TSL
February 8, 2012 4:55 AM
|
Understanding the factors driving pathogen emergence and re-emergence is a major challenge, particularly in agriculture, where the use of resistant plant cultivars imposes strong selective pressures on plant pathogen populations and leads frequently to ‘resistance breakdown’. Presently, durable resistances are only identified after a long period of large-scale cultivation of resistant cultivars. We propose a new predictor of the durability of plant resistance. Because resistance breakdown involves modifications in the avirulence factors of pathogens, we tested for correlations between the evolutionary constraints acting on avirulence factors or their diversity and the durability of the corresponding resistance genes in the case of plant–virus interactions. An analysis performed on 20 virus species–resistance gene combinations revealed that the selective constraints applied on amino acid substitutions in virus avirulence factors correlate with the observed durability of the corresponding resistance genes. On the basis of this result, a model predicting the potential durability of resistance genes as a function of the selective constraints applied on the corresponding avirulence factors is proposed to help breeders to select the most durable resistance genes.

|
Scooped by
Kamoun Lab @ TSL
February 5, 2012 7:37 AM
|
A diverse range of organisms interact with and alter plant immunity through secreted molecules known as effectors. Although a large number of effector molecules have been discovered in the genomes of a range of plant-associated organisms, relatively little is known about the activities and host targets of these effectors. This Symposium will focus on a mechanistic understanding of this process, covering a broad spectrum of plant interactions from divergent taxa including: bacteria, fungi, oomycetes, nematodes, insects and parasitic plants. This will help to illustrate the diversity of evolutionary strategies that microbes and other organisms have evolved that result in both positive and negative alterations to plants. By bringing together top scientists in this field we will provide an opportunity to compare the different approaches being undertaken to unravel the biochemical activities of the effectors. The Symposium will undoubtedly stimulate discussion as well as an exchange of ideas and research strategies.

|
Scooped by
Kamoun Lab @ TSL
February 2, 2012 12:16 PM
|
ToxA is a proteinaceous necrotrophic effector produced by Stagonospora nodorum and Pyrenophora tritici-repentis. In this study, all eight mature isoforms of the ToxA protein were purified and compared. Circular dichroism spectra indicated that all isoforms were structurally intact and had indistinguishable secondary structural features. ToxA isoforms were infiltrated into wheat lines that carry the sensitivity gene Tsn1. It was observed that different wheat lines carrying identical Tsn1 alleles varied in sensitivity to ToxA. All ToxA isoforms induced necrosis when introduced into any Tsn1 wheat line, but we observed quantitative variation in effector activity, with the least active version found in isolates of P. tritici-repentis. Pathogen sporulation increased with higher doses of ToxA. The isoforms that induced the most rapid necrosis also induced the most sporulation, indicating that pathogen fitness is affected by differences in ToxA activity. We show that differences in toxin activity encoded by a single gene can contribute to the quantitative inheritance of necrotrophic virulence. Our findings support the hypothesis that the variation at ToxA results from selection that favors increased toxin activity.

|
Scooped by
Kamoun Lab @ TSL
February 1, 2012 12:21 PM
|
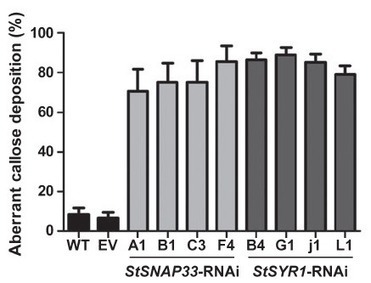
• The oomycete Phytophthora infestans is the causal agent of late blight, the most devastating disease of potato. The importance of vesicle fusion processes and callose deposition for defense of potato against Phytophthora infestans was analyzed.
• Transgenic plants were generated, which express RNA interference constructs targeted against plasma membrane-localized SYNTAXIN-RELATED 1 (StSYR1) and SOLUBLE N-ETHYLMALEIMIDE-SENSITIVE FACTOR ADAPTOR PROTEIN 33 (StSNAP33), the potato homologs of Arabidopsis AtSYP121 and AtSNAP33, respectively.
• Phenotypically, transgenic plants grew normally, but showed spontaneous necrosis and chlorosis formation at later stages. In response to infection with Phytophthora infestans, increased resistance of StSYR1-RNAi plants, but not StSNAP33-RNAi plants, was observed. This increased resistance correlated with the constitutive accumulation of salicylic acid and PR1 transcripts. Aberrant callose deposition in Phytophthora infestans-infected StSYR1-RNAi plants coincided with decreased papilla formation at penetration sites. Resistance against the necrotrophic fungus Botrytis cinerea was not significantly altered. Infiltration experiments with bacterial solutions of Agrobacterium tumefaciens and Escherichia coli revealed a hypersensitive phenotype of both types of RNAi lines.
• The enhanced defense status and the reduced growth of Phytophthora infestans on StSYR1-RNAi plants suggest an involvement of syntaxins in secretory defense responses of potato and, in particular, in the formation of callose-containing papillae.

|
Scooped by
Kamoun Lab @ TSL
January 26, 2012 3:38 PM
|
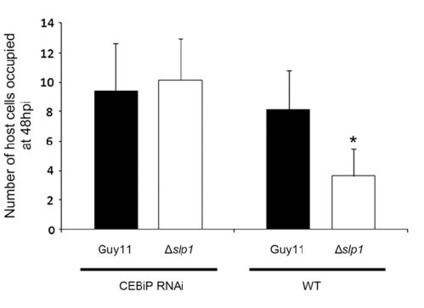
Plants use pattern recognition receptors to defend themselves from microbial pathogens. These receptors recognize pathogen-associated molecular patterns (PAMPs) and activate signaling pathways that lead to immunity. In rice (Oryza sativa), the chitin elicitor binding protein (CEBiP) recognizes chitin oligosaccharides released from the cell walls of fungal pathogens. Here, we show that the rice blast fungus Magnaporthe oryzae overcomes this first line of plant defense by secreting an effector protein, Secreted LysM Protein1 (Slp1), during invasion of new rice cells. We demonstrate that Slp1 accumulates at the interface between the fungal cell wall and the rice plasma membrane, can bind to chitin, and is able to suppress chitin-induced plant immune responses, including generation of reactive oxygen species and plant defense gene expression. Furthermore, we show that Slp1 competes with CEBiP for binding of chitin oligosaccharides. Slp1 is required by M. oryzae for full virulence and exerts a significant effect on tissue invasion and disease lesion expansion. By contrast, gene silencing of CEBiP in rice allows M. oryzae to cause rice blast disease in the absence of Slp1. We propose that Slp1 sequesters chitin oligosaccharides to prevent PAMP-triggered immunity in rice, thereby facilitating rapid spread of the fungus within host tissue.

|
Scooped by
Kamoun Lab @ TSL
January 17, 2012 11:36 AM
|
Late blight (LB) caused by the oomycete Phytophthora infestans, is a major disease of potato and tomato worldwide and can cause up to 100% yield losses. The devastating economic impact of this disease intensified the related pathology and genetics research since the occurrence of Irish famine in 1840s, with a side gain of major scientific discoveries. For example, many of the crucial steps involved in LB defense response in host plants have been elucidated through the use of modern cytological and molecular biology techniques. Also, genetic and biochemical studies have revealed differences between oomycetes and pathogenic fungi, which has led to more selective use of chemicals for LB control. Furthermore, the discovery of P. infestans two mating types and the resultant generation of more aggressive lineages by sexual recombination stresses the need for an integrated and sustainable approach to LB control. These measures would include the use of cultural practices, selective fungicide applications, and genetic resistance. In potato at least a dozen major resistance genes and several quantitative trait loci (QTLs) for LB resistance have been identified, and most modern cultivars have been bred with one or more resistance genes. In tomato, though most commercial cultivars are susceptible to LB, a few major resistance genes and several QTLs have been identified and several breeding programs around the world are developing breeding lines and commercial cultivars with LB resistance. Most recently, a few fresh-market tomato hybrid cultivars with LB resistance were released by the North Carolina State University Tomato Breeding Program in the United States. There is, however, an insufficient number of potato and tomato cultivars with LB resistance, resulting in continued expensive as well as the hazardous and increasingly ineffective use of chemicals for disease control. In an era when both host plants and P. infestans genomes are sequenced and considerable genomic information is available, it is not unexpected that a more sustainable solution to controlling LB is on the horizon. In this review, we summarize the recent achievements in better understanding of the P. infestans pathogenesis, host-pathogen interactions, and the progress made in developing genetic resistance in potato and tomato.

|
Rescooped by
Kamoun Lab @ TSL
from Publications
January 13, 2012 2:55 AM
|
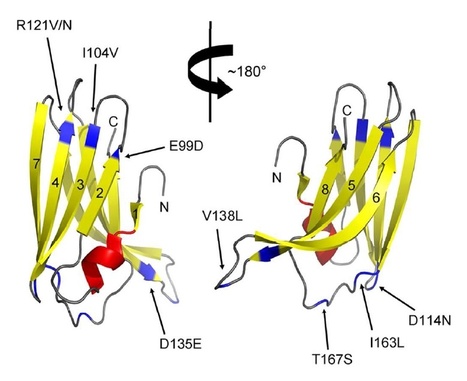
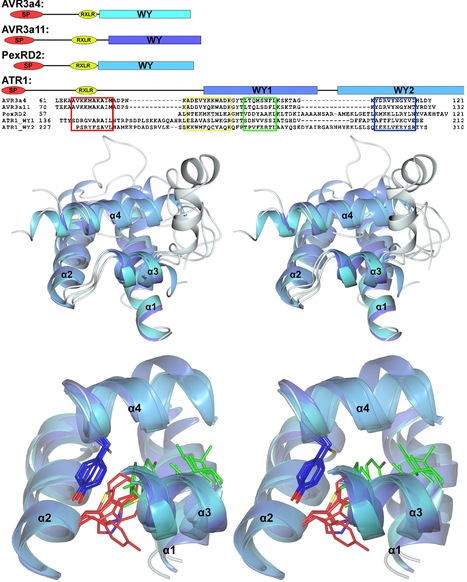
Our laboratories have employed structural biology to investigate the molecular basis of RXLR effector function. A total of four structures have recently been published, those of AVR3a4 and AVR3a11 (paralogues from Phytophthora capsici), PexRD2 (from P. infestans), and ATR1 (from H. arabidopsidis) [21]–[23]. Each publication focused on a different aspect of structure/function analysis including phospholipid binding, protein folding, and effector recognition by the host. The studies of Boutemy et al. and Chou et al. independently described the structural homology of AVR3a11 and a domain of ATR1, respectively, to the cyanobacterial four-helix bundle protein KaiA [24]. This strongly implied they would also be structurally related to each other. This is unexpected, as these Phytophthora and H. arabidopsidis effectors do not share any significant sequence similarity: the conservation was only apparent after the structures were determined and compared.
|

 Your new post is loading...
Your new post is loading...