Your new post is loading...
 Your new post is loading...

|
Rescooped by
Kamoun Lab @ TSL
from Fungal|Oomycete Biology
April 6, 2012 12:42 PM
|
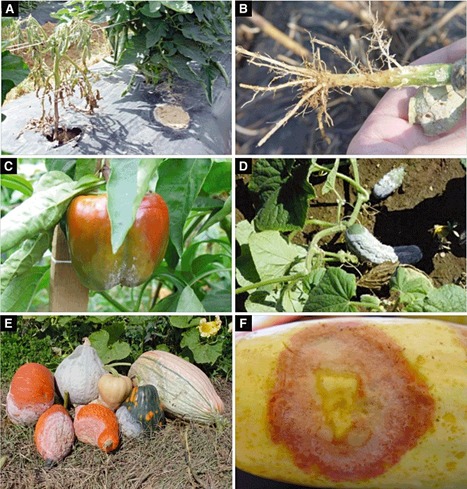
Phytophthora capsici is a highly dynamic and destructive pathogen of vegetables. It attacks all cucurbits, pepper, tomato and eggplant, and, more recently, snap and lima beans. The disease incidence and severity have increased significantly in recent decades and the molecular resources to study this pathogen are growing and now include a reference genome. At the population level, the epidemiology varies according to the geographical location, with populations in South America dominated by clonal reproduction, and populations in the USA and South Africa composed of many unique genotypes in which sexual reproduction is common. Just as the impact of crop loss as a result of P. capsici has increased in recent decades, there has been a similar increase in the development of new tools and resources to study this devastating pathogen. Phytophthora capsici presents an attractive model for understanding broad-host-range oomycetes, the impact of sexual recombination in field populations and the basic mechanisms of Phytophthora virulence.
Via Alejandro Rojas

|
Scooped by
Kamoun Lab @ TSL
April 6, 2012 12:07 PM
|
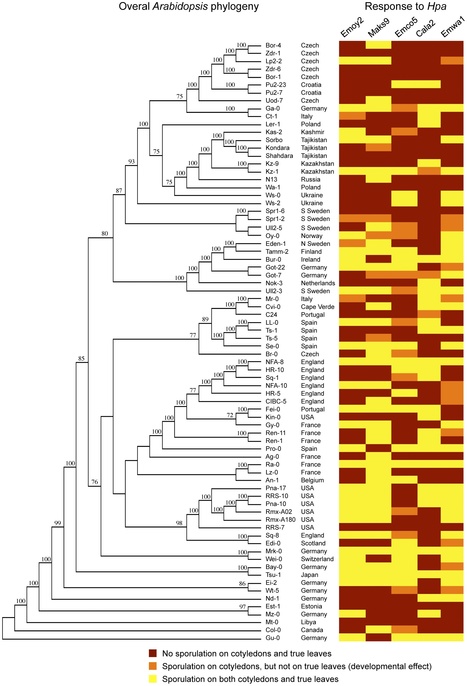
Interactions between Arabidopsis thaliana and its native obligate oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) represent a model system to study evolution of natural variation in a host/pathogen interaction. Both Arabidopsis and Hpa genomes are sequenced and collections of different sub-species are available. We analyzed ~400 interactions between different Arabidopsis accessions and five strains of Hpa. We examined the pathogen's overall ability to reproduce on a given host, and performed detailed cytological staining to assay for pathogen growth and hypersensitive cell death response in the host. We demonstrate that intermediate levels of resistance are prevalent among Arabidopsis populations and correlate strongly with host developmental stage. In addition to looking at plant responses to challenge by whole pathogen inoculations, we investigated the Arabidopsis resistance attributed to recognition of the individual Hpa effectors, ATR1 and ATR13. Our results suggest that recognition of these effectors is evolutionarily dynamic and does not form a single clade in overall Arabidopsis phylogeny for either effector. Furthermore, we show that the ultimate outcome of the interactions can be modified by the pathogen, despite a defined gene-for-gene resistance in the host. These data indicate that the outcome of disease and disease resistance depends on genome-for-genome interactions between the host and its pathogen, rather than single gene pairs as thought previously.

|
Scooped by
Kamoun Lab @ TSL
April 4, 2012 10:57 AM
|
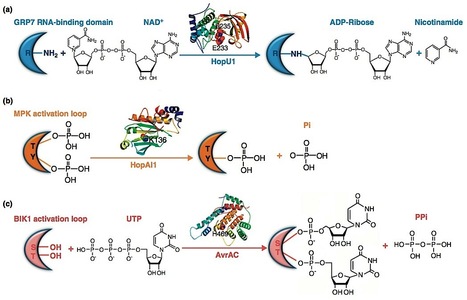
Effectors secreted by the bacterial type III system play a central role in the interaction between Gram-negative bacterial pathogens and their host plants. Recent advances in the effector studies have helped cementing several key concepts concerning bacterial pathogenesis, plant immunity, and plant–pathogen co-evolution. Type III effectors use a variety of biochemical mechanisms to target specific host proteins or DNA for pathogenesis. The identifications of their host targets led to the identification of novel components of plant innate immune system. Key modules of plant immune signaling pathways such as immune receptor complexes and MAPK cascades have emerged as a major battle ground for host–pathogen adaptation. These modules are attacked by multiple type III effectors, and some components of these modules have evolved to actively sense the effectors and trigger immunity.

|
Scooped by
Kamoun Lab @ TSL
March 31, 2012 4:33 AM
|
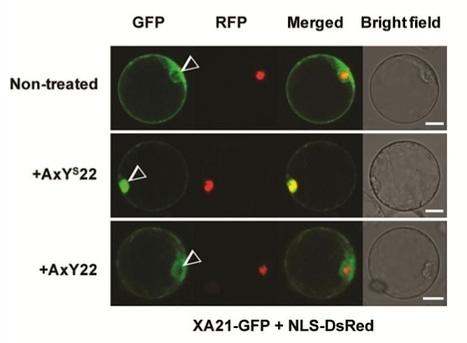
The rice XA21 receptor confers immunity to the Gram-negative bacterial pathogen, Xanthomonas oryzae pv. oryzae (Xoo) upon recognition of the conserved microbial signature AxYS22. Here, we demonstrate that the intracellular kinase domain of XA21 translocates to the nucleus upon AxYS22-mediated perception and that this translocation event is required for XA21-mediated immunity.

|
Scooped by
Kamoun Lab @ TSL
March 28, 2012 5:43 AM
|
Ustilago maydis is a biotrophic pathogen causing maize (Zea mays) smut disease. Transcriptome profiling of infected maize plants indicated that a gene encoding a putative cystatin (CC9) is induced upon penetration by U. maydis wild type. By contrast, cc9 is not induced after infection with the U. maydis effector mutant Δpep1, which elicits massive plant defenses. Silencing of cc9 resulted in a strongly induced maize defense gene expression and a hypersensitive response to U. maydis wild-type infection. Consequently, fungal colonization was strongly reduced in cc9-silenced plants, while recombinant CC9 prevented salicylic acid (SA)–induced defenses. Protease activity profiling revealed a strong induction of maize Cys proteases in SA-treated leaves, which could be inhibited by addition of CC9. Transgenic maize plants overexpressing cc9-mCherry showed an apoplastic localization of CC9. The transgenic plants showed a block in Cys protease activity and SA-dependent gene expression. Moreover, activated apoplastic Cys proteases induced SA-associated defense gene expression in naïve plants, which could be suppressed by CC9. We show that apoplastic Cys proteases play a pivotal role in maize defense signaling. Moreover, we identified cystatin CC9 as a novel compatibility factor that suppresses Cys protease activity to allow biotrophic interaction of maize with the fungal pathogen U. maydis.

|
Scooped by
Kamoun Lab @ TSL
March 26, 2012 7:13 AM
|
A Triticum isolate (pathogenic on wheat) of Magnaporthe oryzae was crossed with an Oryza isolate (pathogenic on rice) to elucidate mechanisms of their parasitic specificity on wheat. When the pathogenicity of their F 1 cultures (hybrids between a Triticum isolate and an Oryza isolate) was tested on wheat, avirulent and virulent cultures segregated in a 7:1 ratio. This result suggests that three loci are involved in avirulence of the Oryza isolate on wheat. One of the three loci conditioned papilla formation, whereas the others conditioned the hypersensitive reaction. Allelism tests revealed that the locus conditioning papilla formation is Pwt2 while one of the two loci conditioning the hypersensitive reaction is Pwt1. The other locus conditioning the hypersensitive reaction was different from any other known loci and, therefore, was designated as Pwt5.

|
Scooped by
Kamoun Lab @ TSL
March 16, 2012 3:56 AM
|
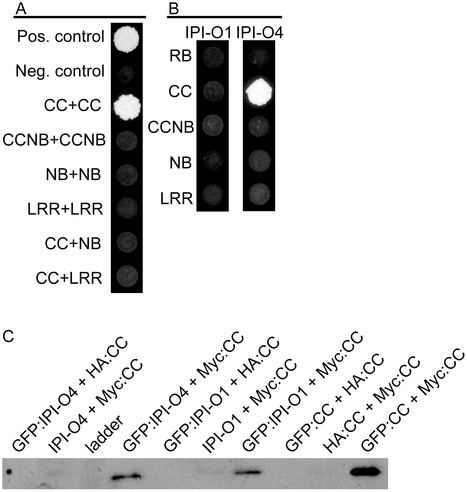
Despite intensive breeding efforts, potato late blight, caused by the oomycete pathogen Phytophthora infestans, remains a threat to potato production worldwide because newly evolved pathogen strains have consistently overcome major resistance genes. The potato RB gene, derived from the wild species Solanum bulbocastanum, confers resistance to most P. infestans strains through recognition of members of the pathogen effector family IPI-O. While the majority of IPI-O proteins are recognized by RB to elicit resistance (e.g. IPI-O1, IPI-O2), some family members are able to elude detection (e.g. IPI-O4). In addition, IPI-O4 blocks recognition of IPI-O1, leading to inactivation of RB-mediated programmed cell death. Here, we report results that elucidate molecular mechanisms governing resistance elicitation or suppression of RB by IPI-O. Our data indicate self-association of the RB coiled coil (CC) domain as well as a physical interaction between this domain and the effectors IPI-O4 and IPI-O1. We identified four amino acids within IPI-O that are critical for interaction with the RB CC domain and one of these amino acids, at position 129, determines hypersensitive response (HR) elicitation in planta. IPI-O1 mutant L129P fails to induce HR in presence of RB while IPI-O4 P129L gains the ability to induce an HR. Like IPI-O4, IPI-O1 L129P is also able to suppress the HR mediated by RB, indicating a critical step in the evolution of this gene family. Our results point to a model in which IPI-O effectors can affect RB function through interaction with the RB CC domain.

|
Scooped by
Kamoun Lab @ TSL
March 14, 2012 1:01 PM
|
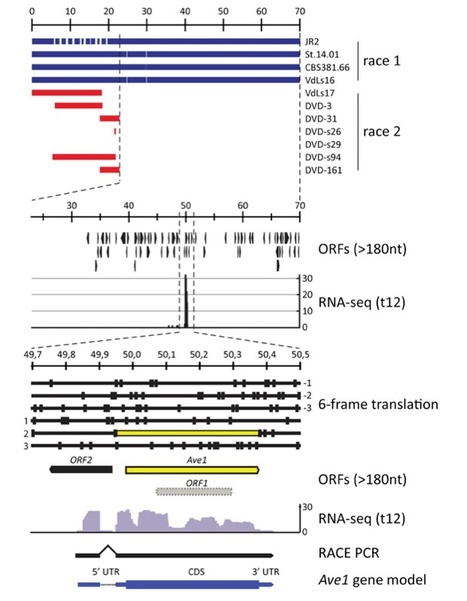
Fungal plant pathogens secrete effector molecules to establish disease on their hosts, and plants in turn use immune receptors to try to intercept these effectors. The tomato immune receptor Ve1 governs resistance to race 1 strains of the soil-borne vascular wilt fungi Verticillium dahliae and Verticillium albo-atrum, but the corresponding Verticillium effector remained unknown thus far. By high-throughput population genome sequencing, a single 50-Kb sequence stretch was identified that only occurs in race 1 strains, and subsequent transcriptome sequencing of Verticillium-infected Nicotiana benthamiana plants revealed only a single highly expressed ORF in this region, designated Ave1 (for Avirulence on Ve1 tomato). Functional analyses confirmed that Ave1 activates Ve1-mediated resistance and demonstrated that Ave1 markedly contributes to fungal virulence, not only on tomato but also on Arabidopsis. Interestingly, Ave1 is homologous to a widespread family of plant natriuretic peptides. Besides plants, homologous proteins were only found in the bacterial plant pathogen Xanthomonas axonopodis and the plant pathogenic fungi Colletotrichum higginsianum, Cercospora beticola, and Fusarium oxysporum f. sp. lycopersici. The distribution of Ave1 homologs, coincident with the presence of Ave1 within a flexible genomic region, strongly suggests that Verticillium acquired Ave1 from plants through horizontal gene transfer. Remarkably, by transient expression we show that also the Ave1 homologs from F. oxysporum and C. beticola can activate Ve1-mediated resistance. In line with this observation, Ve1 was found to mediate resistance toward F. oxysporum in tomato, showing that this immune receptor is involved in resistance against multiple fungal pathogens.

|
Scooped by
Kamoun Lab @ TSL
March 9, 2012 12:58 AM
|
Abstract Submissions Process for the XV International Congress Ends April 17. Don’t miss this opportunity to showcase your research at the XV International Congress on MPMI in Kyoto, Japan, July 30–August 2. Other key deadlines are also approaching, including the deadline for Travel Award Applications (April 17) and Visa Support (April 22). Complete details are available on the newly redesigned congress website http://mpmi2011.umin.jp/. Do you have colleagues that want to attend? Tell them to join now and receive special savings on membership!

|
Scooped by
Kamoun Lab @ TSL
March 5, 2012 1:04 PM
|
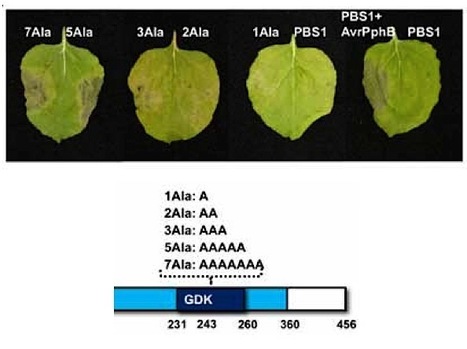
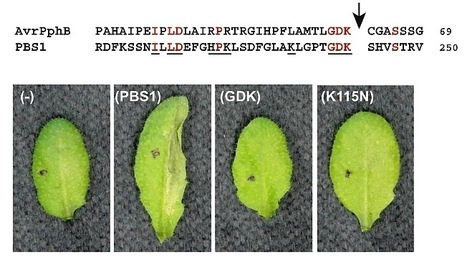
Nucleotide binding-leucine rich repeat (NB-LRR) proteins function as intracellular receptors for the detection of pathogens in both plants and animals. Despite their central role in innate immunity, the molecular mechanisms that govern NB-LRR activation are poorly understood. The Arabidopsis NB-LRR protein RPS5 detects the presence of the Pseudomonas syringae effector protein AvrPphB by monitoring the status of the Arabidopsis protein kinase PBS1. AvrPphB is a cysteine protease that targets PBS1 for cleavage at a single site within the activation loop of PBS1. Using a transient expression system in the plant Nicotiana benthamiana and stable transgenic Arabidopsis plants we found that both PBS1 cleavage products are required to activate RPS5 and can do so in the absence of AvrPphB. We also found, however, that the requirement for cleavage of PBS1 could be bypassed simply by inserting five amino acids at the PBS1 cleavage site, which is located at the apex of the activation loop of PBS1. Activation of RPS5 did not require PBS1kinase function, thus RPS5 appears to sense a subtle conformational change in PBS1, rather than cleavage. This finding suggests that NB-LRR proteins may function as fine-tuned sensors of alterations in the structures of effector targets.

|
Scooped by
Kamoun Lab @ TSL
March 3, 2012 12:08 PM
|
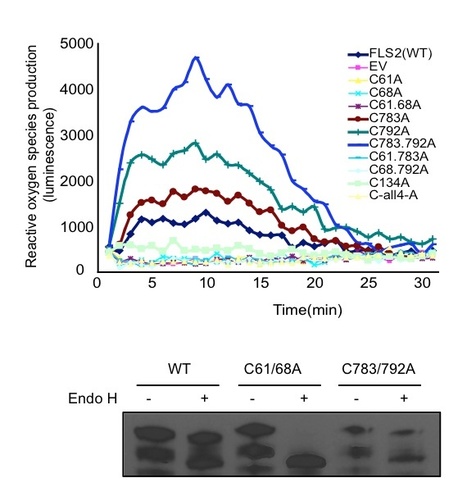
FLAGELLIN SENSING2 (FLS2) is a transmembrane receptor kinase that activates antimicrobial defense responses upon binding of bacterial flagellin or the flagellin-derived peptide flg22. We find that some Arabidopsis thaliana FLS2 is present in FLS2-FLS2 complexes before and after plant exposure to flg22. flg22 binding capability is not required for FLS2-FLS2 association. Cys pairs flank the extracellular leucine rich repeat (LRR) domain in FLS2 and many other LRR receptors, and we find that the Cys pair N-terminal to the FLS2 LRR is required for normal processing, stability, and function, possibly due to undescribed endoplasmic reticulum quality control mechanisms. By contrast, disruption of the membrane-proximal Cys pair does not block FLS2 function, instead increasing responsiveness to flg22, as indicated by a stronger oxidative burst. There was no evidence for intermolecular FLS2-FLS2 disulfide bridges. Truncated FLS2 containing only the intracellular domain associates with full-length FLS2 and exerts a dominant-negative effect on wild-type FLS2 function that is dependent on expression level but independent of the protein kinase capacity of the truncated protein. FLS2 is insensitive to disruption of multiple N-glycosylation sites, in contrast with the related receptor EF-Tu RECEPTOR that can be rendered nonfunctional by disruption of single glycosylation sites. These and additional findings more precisely define the molecular mechanisms of FLS2 receptor function.

|
Scooped by
Kamoun Lab @ TSL
February 24, 2012 4:43 AM
|
Cover Girl: The white rust oomycete Albugo laibachii

|
Scooped by
Kamoun Lab @ TSL
February 22, 2012 3:55 AM
|
The genetically tractable endophytic fungus Piriformospora indica is able to colonize the root cortex of a great variety of different plant species with beneficial effects to its hosts and it represents a suitable model system to study symbiotic interactions. Recent cytological studies in barley and Arabidopsis showed that upon penetration of the root, P. indica establishes a biotrophic interaction during which fungal cells are encased by the host plasma membrane. Large-scale transcriptional analyses of fungal and plant responses have shown that perturbance of plant hormone homeostasis and secretion of fungal lectins and other small proteins (effectors) may be involved in the evasion and suppression of host defenses at these early colonization steps. At later stages P. indica is found more often in moribund host cells where it secretes a large variety of hydrolytic enzymes that degrade proteins. This strategy of colonizing plants is reminiscent of that of hemibiotrophic fungi, although a defined shift to necrotrophy with massive host cell death is missing. Instead, the association with the plant root leads to beneficial effects for the host such as growth promotion, increased resistance to root as well as leaf pathogens, and increased tolerance to abiotic stresses. This review describes current advances in understanding the components of P. indica endophytic lifestyle from molecular and genomic analyses.
|

|
Scooped by
Kamoun Lab @ TSL
April 6, 2012 12:11 PM
|
The model plant Arabidopsis thaliana exhibits extensive natural variation in resistance to parasites. Immunity is often conferred by resistance (R) genes that permit recognition of specific races of a disease. The number of such R genes and their distribution are poorly understood. In this study, we investigated the basis for resistance to the downy mildew agent Hyaloperonospora arabidopsidis ex parasitica (Hpa) in a global sample of A. thaliana. We implemented a combined genome-wide mapping of resistance using populations of recombinant inbred lines and a collection of wild A. thaliana accessions. We tested the interaction between 96 host genotypes collected worldwide and five strains of Hpa. Then, a fraction of the species-wide resistance was genetically dissected using six recently constructed populations of recombinant inbred lines. We found that resistance is usually governed by single dominant R genes that are concentrated in four genomic regions only. We show that association genetics of resistance to diseases such as downy mildew enables increased mapping resolution from quantitative trait loci interval to candidate gene level. Association patterns in quantitative trait loci intervals indicate that the pool of A. thaliana resistance sources against the tested Hpa isolates may be predominantly confined to six RPP (Resistance to Hpa) loci isolated in previous studies. Our results suggest that combining association and linkage mapping could accelerate resistance gene discovery in plants.

|
Rescooped by
Kamoun Lab @ TSL
from Publications
April 5, 2012 4:04 AM
|
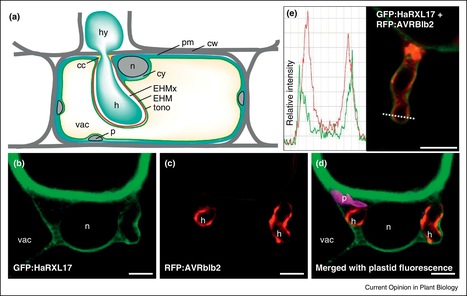
Plant pathogenic oomycetes secrete a diverse repertoire of effector proteins that modulate host innate immunity and enable parasitic infection. Understanding how effectors evolve, translocate and traffic inside host cells, and perturb host processes are major themes in the study of oomycete–plant interactions. The last year has seen important progress in the study of oomycete effectors with, notably, the elucidation of the 3D structures of five RXLR effectors, and novel insights into how cytoplasmic effectors subvert host cells. In this review, we discuss these and other recent advances and highlight the most important open questions in oomycete effector biology.

|
Scooped by
Kamoun Lab @ TSL
March 31, 2012 9:24 PM
|
LA nature toute prodigue qu'elle est nous fournit peu de Plantes d'un aussi grand usàge que le Safran. Ses fleurs font agréables à la vue & à l'odorat. Son pistile est consideré comme une chose précieuse. Il entre dans les apprefts de cuisine; il fournit aux Teinturiers une très belle couleur; les Médecins l'employent très utilements dans plusieurs maladies: sa Fanne même & ses pétales servent dans le pays où on le cultive, à faire du Fourrage pour les bestiaux. Mais semblable en cela aux Plantes les plus précieuses, celle-ci est tendre & délicate, & ne peut être conservée que par des soins proportionnés à ses usages. C'est pourquoy quelque précaution que les habitans du Gaslinois qui la cultivent prennent pour sa conservation , elle ne laisse pas d'être attaquée de plusieurs maladies, qui toutes tendent à la détruire. De toutes celles ausquelles cette Plante est sujette, il n'y en a point de plus dangereuse, ni qui lui soit plus nuisible que celle que les habitans du pays appellent la Mort. Et j'ai été surpris des desordres que cause cette maladie dans les endroits qui ont le malheur d'en être affligés. Et qui ne le seroit pas en effet, de voir qu'une Plante attaquée d'une maladie devient meurtrière des autres de son espece? En avoit-on jusqu'ici remarqué de contagieuses Epidemiques dans les Plantes? Celle qui attaque l'Oignon du Safran est cependant de cette nature,. puisque semblable à la peste des animaux , elle gâte les Oignons voisins , & bientost l'extrémité du champ le sentiroit de la contagion si l'on n'empêchoit la communication par une profonde tranchée qu'il est essentiel de faire dès le commencement du Printemps; parce que la Mort qui fait beaucoup de progrès dans cette saison, n'en fait presque point dans les autres; circonstance digne de remarque, dans la saison où les Plantes paroissent le plus en état de résister à la contagion, elles y succombent, & périssent en plus grand nombre.

|
Scooped by
Kamoun Lab @ TSL
March 30, 2012 4:16 AM
|
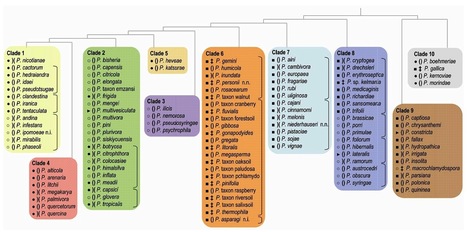
Plant diseases caused by Phytophthora species will remain an ever increasing threat to agriculture and natural ecosystems. Phytophthora literally means plant destroyer, a name coined in the 19th century by Anton de Bary when he investigated the potato disease that set the stage for the Great Irish Famine. Phytophthora infestans, the causal agent of potato late blight, was the first species in a genus that at present has over 100 recognized members. In the last decade, the number of recognized Phytophthora species has nearly doubled and new species are added almost on a monthly basis. Here we present an overview of the 10 clades that are currently distinguished within the genus Phytophthora with special emphasis on new species that have been described since 1996 when Erwin and Ribeiro published the valuable monograph ‘Phytophthora diseases worldwide’ (35).

|
Scooped by
Kamoun Lab @ TSL
March 26, 2012 10:49 AM
|
The SolRgene database consists of 1.062 accessions, and 12.584 genotypes, that were explored for R genes to Phytophthora infestans. Potato (S. tuberosum) is the third most important food crop word-wide, but a major constraint to potato production is that potato cultivars are susceptible to many pathogens. In the tuber-bearing Solanum species, a wealth of R genes is present, which is ready to be exploited. From the Solanum section Petota that comprises many species, we used more than 1,000 accessions that comprise all phylogenetic groups and geographic origins, and form the basis of SolRgene. A core collection of the SolRgene database is maintained in vitro, and available for research purposes.

|
Scooped by
Kamoun Lab @ TSL
March 19, 2012 3:36 AM
|
Some rust fungi are able to change the morphology of the plants they parasitise. One example of this is Uromyces pisi, which can parasitise the plant Euphorbia cyparissias (Cypress Spurge). In this case the rust fungus is able to cause the plant to produce pseudoflowers. These pseudoflowers are rosettes of leaves which are covered with fungal gametes in sugary nectar of fungal origin. The rust’s gametes are on separate and distinct mycelia (they are heterothallic). It is thought that this could be to entice insects towards the fungus in order to cross fertilize their mating types, and that the fungi depends on the insects to carry out sexual reproduction. This theory was tested by Pfunder and Roy in 1999 by a simple insect exclusion experiment (e). The infected plants were enclosed in a fine mesh gauze to prevent the entry of insects. With insects excluded from the infected plant, they concluded that insect pseudoflower visitations were required for the reproduction of U. pisi.

|
Scooped by
Kamoun Lab @ TSL
March 15, 2012 5:54 AM
|
Potato defends against Phytophthora infestans infection by R-gene-based qualitative resistance as well as a quantitative field resistance. R genes are renowned to be rapidly overcome by this oomycete, and potato cultivars with a decent and durable resistance to current P. infestans populations are hardly available. However, potato cultivar Sarpo Mira has retained resistance in the field over several years. We dissected the resistance of cultivar Sarpo Mira in a segregating population by matching the responses to P. infestans RXLR effectors with race-specific resistance to differential strains. The resistance is based on the combination of four pyramided qualitative R genes and a quantitative R gene that was associated with field resistance. The qualitative R genes include R3a, R3b, R4 and the newly identified Rpi-Smira1. The qualitative resistances matched responses to AVR3a, AVR3b, AVR4 and AVRSmira1 RXLR effectors and were overcome by particular P. infestans strains. The quantitative resistance was determined to be conferred by a novel gene Rpi-Smira2. It was only detected under field conditions and was associated with responses to the RXLR effector AvrSmira2. We foresee that effector-based resistance breeding will facilitate selecting and combining qualitative and quantitative resistances that may lead to a more durable resistance to late blight.

|
Scooped by
Kamoun Lab @ TSL
March 13, 2012 5:24 AM
|
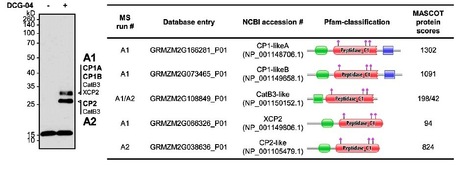
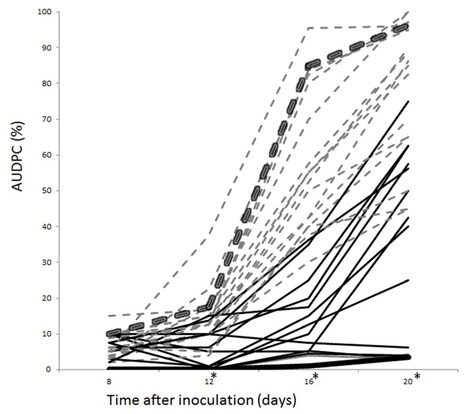
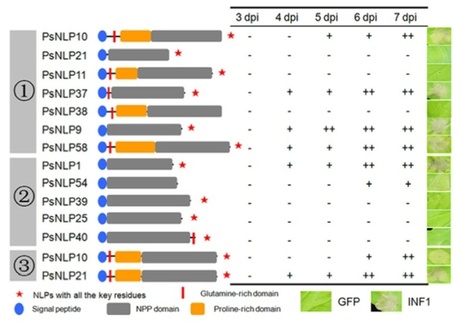
Necrosis and ethylene inducing-like proteins (NLP) are widely distributed in eukaryotic and prokaryotic plant pathogens and are considered to be important virulence factors. We identified a total of 70 potential Phytophthora sojae NLP genes but 37 were designated as pseudogenes. Sequence alignment of the remaining 33 NLPs delineated six groups. Three of these groups include proteins with an intact hepta-peptide (GHRHDWE) motif, which is important for necrosis inducing acitivity, whereas the motif is not conserved in the other groups. A total of 19 representative NLP genes were assessed for necrosis-inducing activity by heterologous expression in Nicotiana benthamiana. Surprisingly, only 8 genes trigger cell death. The expression of the NLP genes in P. sojae was examined, distinguishing 20 expressed and 13 non-expressed NLP genes. Real-time RT-PCR results indicate that most NLPs are highly expressed during cyst germination and infection stages. Amino acid substitution ratios (Ka/Ks) of 33 NLP sequences from four distinct P. sojae strains resulted in identification of positive selection sites in a distinct NLP group. Overall our study indicates that expansion and pseudogenization of the P. sojae NLP family results from an ongoing birth-and-death process, and that varying patterns of expression, necrosis-inducing activity, and positive selection suggest that NLPs have diversified in function.

|
Scooped by
Kamoun Lab @ TSL
March 6, 2012 3:11 PM
|
The aim of this review was to survey all fungal pathologists with an association with the journal Molecular Plant Pathology and ask them to nominate which fungal pathogens they would place in a ‘Top 10’ based on scientific/economic importance. The survey generated 495 votes from the international community, and resulted in the generation of a Top 10 fungal plant pathogen list for Molecular Plant Pathology. The Top 10 list includes, in rank order, (1) Magnaporthe oryzae; (2) Botrytis cinerea; (3) Puccinia spp.; (4) Fusarium graminearum; (5) Fusarium oxysporum; (6) Blumeria graminis; (7) Mycosphaerella graminicola; (8) Colletotrichum spp.; (9) Ustilago maydis; (10) Melampsora lini, with honourable mentions for fungi just missing out on the Top 10, including Phakopsora pachyrhizi and Rhizoctonia solani. This article presents a short resumé of each fungus in the Top 10 list and its importance, with the intent of initiating discussion and debate amongst the plant mycology community, as well as laying down a bench-mark. It will be interesting to see in future years how perceptions change and what fungi will comprise any future Top 10.

|
Scooped by
Kamoun Lab @ TSL
March 5, 2012 1:22 PM
|
Plant disease-resistance (R) proteins are thought to function as receptors for ligands produced directly or indirectly by pathogen avirulence (Avr) proteins. The biochemical functions of most Avr proteins are unknown, and the mechanisms by which they activate R proteins have not been determined. In Arabidopsis, resistance to Pseudomonas syringae strains expressing AvrPphB requires RPS5, a member of the class of R proteins that have a predicted nucleotide-binding site and leucine-rich repeats, and PBS1, a protein kinase. AvrPphB was found to proteolytically cleave PBS1, and this cleavage was required for RPS5-mediated resistance, which indicates that AvrPphB is detected indirectly via its enzymatic activity.

|
Scooped by
Kamoun Lab @ TSL
March 2, 2012 8:41 AM
|
Plants are engaged in a continuous co-evolutionary struggle for dominance with their pathogens. The outcomes of these interactions are of particular importance to human activities, as they can have dramatic effects on agricultural systems. The recent convergence of molecular studies of plant immunity and pathogen infection strategies is revealing an integrated picture of the plant–pathogen interaction from the perspective of both organisms. Plants have an amazing capacity to recognize pathogens through strategies involving both conserved and variable pathogen elicitors, and pathogens manipulate the defence response through secretion of virulence effector molecules. These insights suggest novel biotechnological approaches to crop protection.

|
Scooped by
Kamoun Lab @ TSL
February 23, 2012 11:24 AM
|
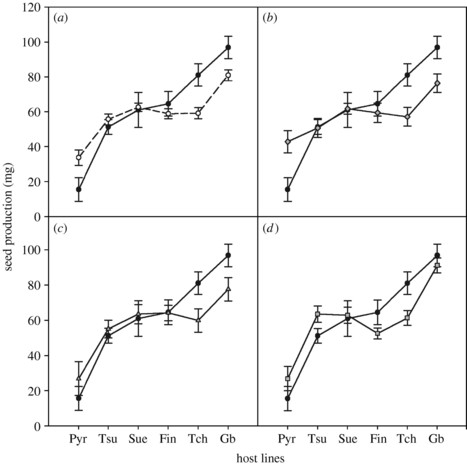
Are parasites always harmful to their hosts? By definition, indeed, but in a few cases and particular environments, hosts experience higher fitness in the presence than in the absence of their parasites. Symbiotic associations form a continuum of interactions, from deleterious to beneficial effects on hosts. In this paper, we investigate the outcome of parasite infection of Arabidopsis thaliana by its natural pathogen Hyaloperonospora arabidopsis. This system exhibits a wide range of parasite impact on host fitness with, surprisingly, deleterious effects on high fecundity hosts and, at the opposite extreme, seemingly beneficial effects on the least fecund one. This phenomenon might result from varying levels of tolerance among host lines and even overcompensation for parasite damage analogous to what can be observed in plant–herbivore systems.
|

 Your new post is loading...
Your new post is loading...