 Your new post is loading...
 Your new post is loading...
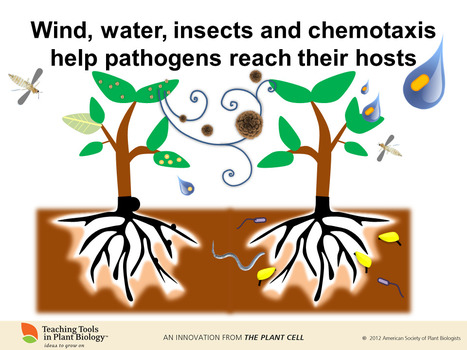
Teaser - coming soon, Plants and Pathogens
Teaching Tools in Plant Biology 22: Plants and Pathogens. Look for it in June. Here's one of our favorite slides.
Via Mary Williams

|
Rescooped by
Kamoun Lab @ TSL
from Plant Pathogenomics
May 24, 2012 8:30 AM
|
Although the oomycetes and fungi are evolutionarily very distantly related, both taxa evolved biotrophy on plant hosts several times independently, giving rise to rust- and mildew-like phenotypes. Differences in host colonization and adaptation may be reflected in genome size and by gain and loss of genes. In this opinion article we combine classical knowledge with recently sequenced pathogen genomes and present new hypotheses about the convergent evolution that led to these two distinct phenotypes in obligate biotrophs.

|
Scooped by
Kamoun Lab @ TSL
May 21, 2012 2:31 PM
|
Due to their potential roles in pathogen defense, genes encoding nucleotide-binding site (NBS) domain have been particularly surveyed in many angiosperm genomes. Two typical classes were found: one is the TIR-NBS-LRR (TNL) class and the other is the CC-NBS-LRR (CNL) class. It is seldom known, however, what kind of NBS-encoding genes are mainly present in other plant groups, especially the most ancient groups of land plants, that is, bryophytes. To fill this gap of knowledge, in this study, we mainly focused on two bryophyte species: the moss Physcomitrella patens and the liverwort Marchantia polymorpha, to survey their NBS-encoding genes. Surprisingly, two novel classes of NBS-encoding genes were discovered. The first novel class is identified from the P. patens genome and a typical member of this class has a protein kinase (PK) domain at the N-terminus and a LRR domain at the C-terminus, forming a complete structure of PK-NBS-LRR (PNL), reminiscent of TNL and CNL classes in angiosperms. The second class is found from the liverwort genome and a typical member of this class possesses an α/β-hydrolase domain at the N-terminus and also a LRR domain at the C-terminus (Hydrolase-NBS-LRR, HNL). Analysis on intron positions and phases also confirmed the novelty of HNL and PNL classes, as reflected by their specific intron locations or phase characteristics. Phylogenetic analysis covering all four classes of NBS-encoding genes revealed a closer relationship among the HNL, PNL and TNL classes, suggesting the CNL class having a more divergent status from the others. The presence of specific introns highlights the chimerical structures of HNL, PNL and TNL genes, and implies their possible origin via exon-shuffling during the quick lineage separation processes of early land plants.

|
Scooped by
Kamoun Lab @ TSL
May 19, 2012 8:32 AM
|
Individual plant and enemy species (or populations) are reciprocally interacting in a way that shapes their traits and evolution. This concept of specificity in plant–herbivore and plant–pathogen interactions is central to this special issue of Trends in Plant Science. Why is it that most herbivores and pathogens attack a minute fraction of the plants or even plant organs available to them? How do plants manage to defend against diverse enemies? Why are plant enemies specialized at all, given that specialization seems to simply limit the number of available hosts? Are most current plant–enemy interactions the result of a coevolutionary history, and can these be manipulated to protect our agricultural crops from pest insects and disease and our ecosystems from invasive species? In this issue we combine perspectives of the plant with that of its enemies, in order to address these questions and focus on the traits that allow for successful plant defense versus successful exploitation of plant tissues. Cover design by Alejandro de León.

|
Scooped by
Kamoun Lab @ TSL
May 16, 2012 10:20 AM
|
Endosymbiotic interactions are characterized by the formation of specialized membrane compartments, by the host in which the microbes are hosted, in an intracellular manner. Two well-studied examples, which are of major agricultural and ecological importance, are the widespread arbuscular mycorrhizal symbiosis and the Rhizobium–legume symbiosis. In both symbioses, the specialized host membrane that surrounds the microbes forms a symbiotic interface, which facilitates the exchange of, for example, nutrients in a controlled manner and, therefore, forms the heart of endosymbiosis. Despite their key importance, the molecular and cellular mechanisms underlying the formation of these membrane interfaces are largely unknown. Recent studies strongly suggest that the Rhizobium–legume symbiosis coopted a signaling pathway, including receptor, from the more ancient arbuscular mycorrhizal symbiosis to form a symbiotic interface. Here, we show that two highly homologous exocytotic vesicle-associated membrane proteins (VAMPs) are required for formation of the symbiotic membrane interface in both interactions. Silencing of these Medicago VAMP72 genes has a minor effect on nonsymbiotic plant development and nodule formation. However, it blocks symbiosome as well as arbuscule formation, whereas root colonization by the microbes is not affected. Identification of these VAMP72s as common symbiotic regulators in exocytotic vesicle trafficking suggests that the ancient exocytotic pathway forming the periarbuscular membrane compartment has also been coopted in the Rhizobium–legume symbiosis.

|
Scooped by
Kamoun Lab @ TSL
May 14, 2012 5:49 PM
|
Epichloae endophytes form mutualistic symbiotic associations with temperate grasses and confer on the host a number of bioprotective benefits through production of fungal secondary metabolites and changed host metabolism. Maintenance of this mutualistic interaction requires that growth of the endophyte within the host is restricted. Recent work has shown that epichloae endophytes grow in the leaves by intercalary division and extension rather than tip growth. This novel pattern of growth enables the fungus to synchronise its growth with that of the host. Reactive oxygen species signalling is required to maintain this pattern of growth. Disruption of components of the NADPH oxidase complex or a MAP kinase, result in a switch from restricted to proliferative growth and a breakdown in the symbiosis. RNAseq analysis of mutant and wild-type associations identifies key fungal and plant genes that define the symbiotic state. Endophyte genes for secondary metabolite biosynthesis are only expressed in the plant and under conditions of restricted growth.

|
Scooped by
Kamoun Lab @ TSL
May 12, 2012 11:32 AM
|
A Westcountry artist whose massive wooden chair once towered over Dartmoor has embarked on another monumental project to highlight the threat posed by a virulent tree disease. Henry Brudenell-Bruce sparked controversy after installing the Giant's Chair near Widecombe-in-the-Moor, which ended with it being removed by the Dartmoor National Park Authority despite its popular appeal. Now he has painted a massive skeletal 70ft high oak tree fluorescent pink at the Delamore Estate arts and sculpture park, near Cornwood, to make visitors aware of the destructive "fungi" Phytophthora ramorum which has been found across Devon and Cornwall. Delamore founder Gavin Dollard said: "Landowners across the South West are felling seven square miles of Japanese larch to combat the tree-killing "fungus" which was discovered in 2002. "Nearly 1,000 acres of infected larch have been removed by the Forestry Commission, while private landowners have been ordered to fell 4,000 acres of timber by March, some 300,000 tons.

|
Rescooped by
Kamoun Lab @ TSL
from TAL effector science
May 10, 2012 6:58 PM
|
Nature Biotech: High-efficiency TALEN-based gene editing produces disease-resistant rice (2012)
http://www.nature.com/nbt/journal/v30/n5/full/nbt.2199.html?WT.ec_id=NBT-201205 Transcription activator–like (TAL) effectors of Xanthomonas oryzae pv. oryzae (Xoo) contribute to pathogen virulence by transcriptionally activating specific rice disease-susceptibility (S) genes. TAL effector nucleases (TALENs)—fusion proteins derived from the DNA recognition repeats of native or customized TAL effectors and the DNA cleavage domains of FokI—have been used to create site-specific gene modifications in plant cells, yeast, animals and even human pluripotent cells. Here, we exploit TALEN technology to edit a specific S gene in rice to thwart the virulence strategy of X. oryzae and thereby engineer heritable genome modifications for resistance to bacterial blight, a devastating disease in a crop that feeds half of the world's population.
Via dromius

|
Scooped by
Kamoun Lab @ TSL
May 10, 2012 6:24 PM
|
The corn smut Ustilago maydis establishes a biotrophic interaction with its host plant maize. This interaction requires efficient suppression of plant immune responses, which is attributed to secreted effector proteins. Previously we identified Pep1 (Protein essential during penetration-1) as a secreted effector with an essential role for U. maydis virulence. pep1 deletion mutants induce strong defense responses leading to an early block in pathogenic development of the fungus. Using cytological and functional assays we show that Pep1 functions as an inhibitor of plant peroxidases. At sites of Δpep1 mutant penetrations, H2O2 strongly accumulated in the cell walls, coinciding with a transcriptional induction of the secreted maize peroxidase POX12. Pep1 protein effectively inhibited the peroxidase driven oxidative burst and thereby suppresses the early immune responses of maize. Moreover, Pep1 directly inhibits peroxidases in vitro in a concentration-dependent manner. Using fluorescence complementation assays, we observed a direct interaction of Pep1 and the maize peroxidase POX12 in vivo. Functional relevance of this interaction was demonstrated by partial complementation of the Δpep1 mutant defect by virus induced gene silencing of maize POX12. We conclude that Pep1 acts as a potent suppressor of early plant defenses by inhibition of peroxidase activity. Thus, it represents a novel strategy for establishing a biotrophic interaction.

|
Scooped by
Kamoun Lab @ TSL
May 3, 2012 4:34 PM
|
Synaptotagmins are calcium sensors that regulate synaptic vesicle exo/endocytosis. Thought to be exclusive to animals, they have recently been characterized in plants. We show that Arabidopsis synaptotagmin SYTA regulates endosome recycling and movement protein (MP)-mediated trafficking of plant virus genomes through plasmodesmata. SYTA localizes to endosomes in plant cells and directly binds the distinct Cabbage leaf curl virus (CaLCuV) and Tobacco mosaic virus (TMV) cell-to-cell movement proteins. In a SYTA knockdown line, CaLCuV systemic infection is delayed, and cell-to-cell spread of TMV and CaLCuV movement proteins is inhibited. A dominant-negative SYTA mutant causes depletion of plasma membrane-derived endosomes, produces large intracellular vesicles attached to plasma membrane, and inhibits cell-to-cell trafficking of TMV and CaLCuV movement proteins, when tested in an Agrobacterium-based leaf expression assay. Our studies show that SYTA regulates endocytosis, and suggest that distinct virus movement proteins transport their cargos to plasmodesmata for cell-to-cell spread via an endocytic recycling pathway.
This is a beautiful resource that illustrates the changing ideas and demographics of phytopathology from 1908 - 2008. Great old photos!
Via Mary Williams

|
Scooped by
Kamoun Lab @ TSL
April 27, 2012 12:15 PM
|
Proteins produced by the large and diverse chitinase gene family are involved in the hydrolyzation of glycosidic bonds in chitin, a polymer of N-acetylglucosamines. In flowering plants, class I chitinases are important pathogenesis-related (PR) proteins, functioning in the determent of herbivory and pathogen attack by acting on insect exoskeletons and fungal cell walls. Within the carnivorous plants, two subclasses of class I chitinases have been identified to play a role in the digestion of prey. Members of these two subclasses, depending on the presence or absence of a C-terminal extension, can be secreted from specialized digestive glands found within the morphologically diverse traps that develop from carnivorous plant leaves. The degree of homology among carnivorous plant class I chitinases and the method by which these enzymes have been adapted for the carnivorous habit has yet to be elucidated. This study focuses on understanding the evolution of carnivory and chitinase genes in one of the major groups of plants that has evolved the carnivorous habit: the Caryophyllales. We recover novel class I chitinase homologs from species of genera Ancistrocladus, Dionaea, Drosera, Nepenthes, and Triphyophyllum, while also confirming the presence of two subclasses of class I chitinases based upon sequence homology and phylogenetic affinity to class I chitinases available from sequenced angiosperm genomes. We further detect residues under positive selection and reveal substitutions specific to carnivorous plant class I chitinases. These substitutions may confer functional differences as indicated by protein structure homology-modeling.

|
Scooped by
Kamoun Lab @ TSL
April 26, 2012 7:41 PM
|
The spores of the plant pathogenic fungus Colletotrichum higginsianum germinate on plant surfaces to produce highly specialized cells called appressoria with thick, darkly melanized cell walls. Long-recognized as organs of attachment and penetration, appressoria also function in the focal delivery of secreted effector proteins (see Kleemann et al., http://tinyurl.com/clcveqz). Transmission electron microscopy reveals a needle-like penetration hypha emerging from a nanoscale pore (200 nm diameter) in the base of the appressorium to puncture the plant cuticle and cell wall. Image Credit: Richard O'Connell, Max Planck Institute for Plant Breeding Research, Cologne http://www.mpipz.mpg.de/23747/project_description
|

|
Scooped by
Kamoun Lab @ TSL
May 24, 2012 9:17 PM
|
Day 1 Updates of #OMGN12 Oomycete Molecular Genetics Network Annual Meeting, Nanjing, China
No twitter access in China so we will use scoop.it for live blogging #OMGN12. Please use the comments option below to post your "tweets".

|
Scooped by
Kamoun Lab @ TSL
May 23, 2012 5:34 AM
|
Oomycete Molecular Genetics Network Annual Meeting, May 26-28, Nanjing, China (2012)

|
Scooped by
Kamoun Lab @ TSL
May 20, 2012 6:55 PM
|
• Plant disease resistance genes (R genes) encode proteins that function to monitor signals indicating pathogenic infection, thus playing a critical role in the plant’s defense system. Although many studies have been performed to explore the functional details of these important genes, their origin and evolutionary history remain unclear.
• In this study, focusing on the largest group of R genes, the nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes, we conducted an extensive genome-wide survey of 38 representative model organisms and obtained insights into the evolutionary stage and timing of NBS-LRR genes.
• Our data show that the two major domains, NBS and LRR, existed before the split of prokaryotes and eukaryotes but their fusion was observed only in land plant lineages. The Toll/interleukin-1 receptor (TIR) class of NBS-LRR genes probably had an earlier origin than its nonTIR counterpart. The similarities of the innate immune systems of plants and animals are likely to have been shaped by convergent evolution after their independent origins.
• Our findings start to unravel the evolutionary history of these important genes from the perspective of comparative genomics and also highlight the important role of reorganizing pre-existing building blocks in generating evolutionary novelties.

|
Scooped by
Kamoun Lab @ TSL
May 17, 2012 7:39 AM
|
Nature: NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants (2012)
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature11162.html Salicylic acid (SA) is a plant immune signal produced after pathogen challenge to induce systemic acquired resistance. It is the only major plant hormone for which the receptor has not been firmly identified. Systemic acquired resistance in Arabidopsis requires the transcription cofactor nonexpresser of PR genes 1 (NPR1), the degradation of which acts as a molecular switch. Here we show that the NPR1 paralogues NPR3 and NPR4 are SA receptors that bind SA with different affinities. NPR3 and NPR4 function as adaptors of the Cullin 3 ubiquitin E3 ligase to mediate NPR1 degradation in an SA-regulated manner. Accordingly, the Arabidopsis npr3 npr4 double mutant accumulates higher levels of NPR1, and is insensitive to induction of systemic acquired resistance. Moreover, this mutant is defective in pathogen effector-triggered programmed cell death and immunity. Our study reveals the mechanism of SA perception in determining cell death and survival in response to pathogen challenge.

|
Scooped by
Kamoun Lab @ TSL
May 16, 2012 5:39 AM
|
Strategy: To defeat an enemy, first discover all you can about him. In this case we must better understand the science of peacock spot. Let us look more closely at the life cycle of the disease-causing fungus - Spilocea oleaginea. The cycle: As with any living organism, a number of distinct stages are involved in the completion of a new generation. Starting with sporulation (at the top of Figure 1) an already-established growth of the fungus releases many millions of tiny (c. 16 µm) conidia (asexually produced spores, cut off from the tip of specialised hyphae). Each spore is capable of generating a new infection. If these spores are to succeed in their task, they must first be dispersed as widely as possible in order to find areas of healthy leaf tissue into which they can grow. When this is done (and other conditions are also met) the spore now germinates, developing a ‘germ tube’ that must penetrate the cuticle of the leaf in order to cause an infection. When infection has occurred, it is only a matter of time before the fungus develops to a point at which it is ready to produce a fresh generation of spores so re-starting the disease cycle.

|
Scooped by
Kamoun Lab @ TSL
May 12, 2012 12:06 PM
|
An entire suburb in New Zealand has been put on 'lockdown' and the nation is on Red Alert after the capture of a feared invader.
Jitters spread throughout the country after the discovery of a single male Queensland fruit fly, described as the world's worst fruit pest. The Queensland fruit fly, Bactrocera tryoni (Froggatt) http://entnemdept.ufl.edu/creatures/fruit/tropical/queensland_fruit_fly.htm

|
Scooped by
Kamoun Lab @ TSL
May 11, 2012 5:56 PM
|
Dyer's woad rust (Puccinia thlaspeos 'strain woad') is used as a pesticide to control the spread of dyer's woad, an invasive weed in the dry open areas of eight western states. Rusts are a group of fungi that infect only plants, and are often very selective about their hosts. Despite extensive testing of related plants, dyer's woad is the only known plant host for this rust. When used according to label directions, pesticide products containing dyer's woad rust present no known risks to humans, non-target plants, wildlife, or the environment. Isatis tinctoria (dyer's woad) http://en.wikipedia.org/wiki/Isatis_tinctoria

|
Scooped by
Kamoun Lab @ TSL
May 10, 2012 6:29 PM
|
To search for virulence effector genes of the rice blast fungus, Magnaporthe oryzae, we carried out a large-scale targeted disruption of genes for 78 putative secreted proteins that are expressed during the early stages of infection of M. oryzae. Disruption of the majority of genes did not affect growth, conidiation, or pathogenicity of M. oryzae. One exception was the gene MC69. The mc69 mutant showed a severe reduction in blast symptoms on rice and barley, indicating the importance of MC69 for pathogenicity of M. oryzae. The mc69 mutant did not exhibit changes in saprophytic growth and conidiation. Microscopic analysis of infection behavior in the mc69 mutant revealed that MC69 is dispensable for appressorium formation. However, mc69 mutant failed to develop invasive hyphae after appressorium formation in rice leaf sheath, indicating a critical role of MC69 in interaction with host plants. MC69 encodes a hypothetical 54 amino acids protein with a signal peptide. Live-cell imaging suggested that fluorescently labeled MC69 was not translocated into rice cytoplasm. Site-directed mutagenesis of two conserved cysteine residues (Cys36 and Cys46) in the mature MC69 impaired function of MC69 without affecting its secretion, suggesting the importance of the disulfide bond in MC69 pathogenicity function. Furthermore, deletion of the MC69 orthologous gene reduced pathogenicity of the cucumber anthracnose fungus Colletotrichum orbiculare on both cucumber and Nicotiana benthamiana leaves. We conclude that MC69 is a secreted pathogenicity protein commonly required for infection of two different plant pathogenic fungi, M. oryzae and C. orbiculare pathogenic on monocot and dicot plants, respectively.
• Root-knot nematodes (RKNs) are obligate endoparasites that maintain a biotrophic relationship with their hosts over a period of several weeks and induce the differentiation of root cells into specialized feeding cells. Nematode effectors synthesized in the oesophageal glands and injected into the plant tissue through the syringe-like stylet certainly play a central role in these processes.
• In a search for nematode effectors, we used comparative genomics on expressed sequence tag (EST) datasets to identify Meloidogyne incognita genes encoding proteins potentially secreted upon the early steps of infection.
• We identified three genes specifically expressed in the oesophageal glands of parasitic juveniles that encode predicted secreted proteins. One of these genes, Mi-EFF1 is a pioneer gene that has no similarity in databases and a predicted nuclear localization signal. We demonstrate that RKNs secrete Mi-EFF1 within the feeding site and show Mi-EFF1 targeting to the nuclei of the feeding cells.
• RKNs were previously shown to secrete proteins in the apoplasm of infected tissues. Our results show that nematodes sedentarily established at the feeding site also deliver proteins within plant cells through their stylet. The protein Mi-EFF1 injected within the feeding cells is targeted at the nuclei where it may manipulate nuclear functions of the host cell.

|
Scooped by
Kamoun Lab @ TSL
May 1, 2012 7:00 AM
|
/via Quaedvlieg et al. Zymoseptoria gen. nov.: a new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia. The Mycosphaerella complex is both poly- and paraphyletic, containing several different families and genera. The genus Mycosphaerella is restricted to species with Ramularia anamorphs, while Septoria is restricted to taxa that cluster with the type species of Septoria, S. cytisi, being closely related to Cercospora in the Mycosphaerellaceae. Species that occur on graminicolous hosts represent an as yet undescribed genus, for which the name Zymoseptoria is proposed. Based on the 28S nrDNA phylogeny derived in this study, Zymoseptoria is shown to cluster apart from Septoria. Morphologically species of Zymoseptoria can also be distinguished by their yeast-like growth in culture, and the formation of different conidial types that are absent in Septoria s.str. Other than the well-known pathogens such as Z. tritici, the causal agent of septoria tritici blotch on wheat, and Z. passerinii, the causal agent of septoria speckled leaf blotch of barley, both for which epitypes are designated, two leaf blotch pathogens are also described on graminicolous hosts from Iran. Zymoseptoria brevis sp. nov. is described from Phalaris minor, and Z. halophila comb. nov. from leaves of Hordeum glaucum. Further collections are now required to elucidate the relative importance, host range and distribution of these species.

|
Rescooped by
Kamoun Lab @ TSL
from Plant Pathogenomics
April 29, 2012 4:59 AM
|
Phytoplasmas are bacterial pathogens of plants that are transmitted by insects. These bacteria uniquely multiply intracellularly in both plants (Plantae) and insects (Animalia). Similarly to bacterial endosymbionts, phytoplasmas have reduced genomes with limited metabolic capabilities. Nonetheless, the chromosomes of many phytoplasmas are rich in repeated DNA consisting of mobile elements. Phytoplasmas produce an arsenal of effectors most of which are encoded on these mobile elements and on plasmids. These effectors target conserved plant transcription factors resulting in witches’ broom and leafy flower symptoms and suppression of plant defense to insect vectors that transmit the phytoplasmas. Future studies of these fascinating microbes will generate a wealth of new knowledge about forces that shape genomes and microbial interactions with multicellular hosts.

|
Scooped by
Kamoun Lab @ TSL
April 27, 2012 6:54 AM
|
The aim of this topic is to solicit transformative solutions to the pest and pathogen pressures faced by smallholder farmers in developing countries. We encourage researchers and entrepreneurs to harness the emerging information and tools in biology and engineering for the goals of agricultural development, to generate ideas that will revolutionize current approaches to crop protection by focusing on the plant, the pests, pathogens, weeds, and/or their interactions. Preliminary data is not required, but proposals should clearly demonstrate how the idea is an innovative leap in progress with the potential to be transformative. Apply before May 15, 2012 http://www.grandchallenges.org/Explorations/Pages/ApplicationInstructions.aspx
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...






