Your new post is loading...
 Your new post is loading...

|
Scooped by
Kamoun Lab @ TSL
June 14, 2012 4:00 PM
|
St. Paul, Minn. — On The Daily Circuit Monday, we're talking about labeling genetically modified food. Recently, we spoke with Les Szabo, research geneticist and acting research leader at the USDA-ARS Cereal Disease Laboratory on The University of Minnesota St. Paul campus. At the lab, scientists study rust on cereal plants like wheat, rye and barley. Wheat rust is very common and it's completely manageable because most of the wheat that's planted in the world has resistance against these kinds of rust. There are even documented cases of rust back to the Roman days, Szabo said.

|
Scooped by
Kamoun Lab @ TSL
June 14, 2012 9:05 AM
|
Nature Biotech: European agricultural policy goes down the tubers (2012)
Crop cultivars developed by BASF include, but are not limited to, potato varieties with altered starch qualities meant for industrial starch production. The company developed several amylopectin-rich cultivars, including Amflora, Amadea and Modena (transgenic for either an antisense or RNA interference construct of granule-bound starch synthase from Solanum tuberosum that suppresses expression of the enzyme). The first of these, Amflora, is one of only two GM products that have been approved for cultivation by the European Union (EU). In addition, BASF has bred another potato cultivar Fortuna, which is transgenic for the Solanum bulbocastanum resistance genes Rpi-blb1 and Rpi-blb2, and shows enhanced resistance to the fungus-like oomycete Phytophthora infestans, which causes potato late blight. http://www.nature.com/nbt/journal/v30/n6/full/nbt.2255.html?WT.ec_id=NBT-201206

|
Rescooped by
Kamoun Lab @ TSL
from Fungal|Oomycete Biology
June 12, 2012 1:29 PM
|
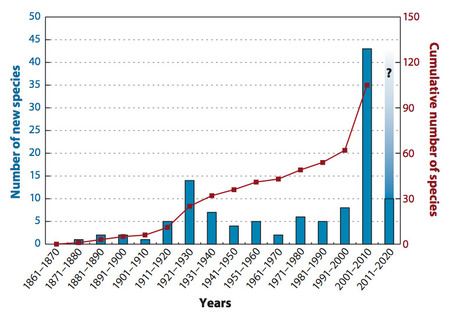
Little is known about indigenous Phytophthora species in natural ecosystems. Increasing evidence, however, suggests that a diverse, trophically complex Phytophthora community is important in many forests. The number of described species has steadily increased, with a dramatic spike in recent years as new species have been split from old and new species have been discovered through exploration of new habitats. Forest soil, streams, and the upper canopies of trees are now being explored for Phytophthoradiversity, and a new appreciation for the ecological amplitude of the genus is emerging. Ten to twenty species are regularly identified in temperate forest surveys. Half or more of this Phytophthoradiversity comes from species described since 2000. Taxa in internal transcribed spacer (ITS) Clade 6 are especially numerous in forest streams and may be saprophytic in this habitat. Three ecological assemblages of forest Phytophthora species are hypothesized: aquatic opportunists, foliar pathogens, and soilborne fine-root and canker pathogens. Aggressive invasive species are associated with all three groups.
Via Alejandro Rojas

|
Scooped by
Kamoun Lab @ TSL
June 12, 2012 4:33 AM
|
Disease resistance involves trade-offs with many other aspects of a plant's phenotype and may cause reduced fitness in terms of growth and reproduction. Likewise, increased virulence to a host plant may lower the fitness of a parasite in other ways. The BSPP 2012 Presidential Conference will bring together leading experts on fitness costs and trade-offs involved in plant disease. It will cover the full range of the subject from molecular biology and physiology to ecology and evolution, as well as the relevance of fitness costs to disease control. Registration will be open in April.

|
Scooped by
Kamoun Lab @ TSL
June 6, 2012 6:41 AM
|
Some thoughts and ideas about effectors. Feel free to add yours on twitter using #effectorwisdom or using the comment option below.

|
Scooped by
Kamoun Lab @ TSL
June 8, 2012 7:12 AM
|
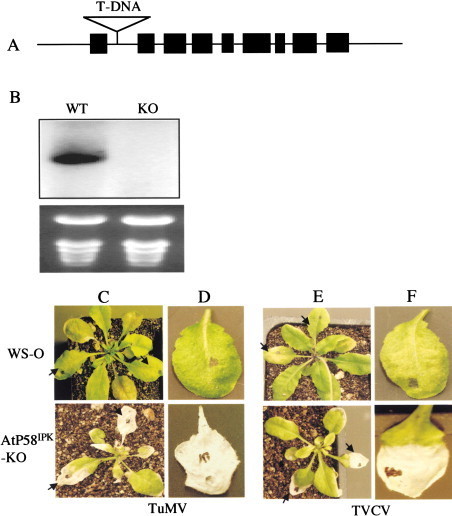
P58IPK is a cellular inhibitor of the mammalian double-stranded RNA-activated protein kinase (PKR). Here we provide evidence for the existence of its homolog in plants and its role in viral infection at the organism level. Viral infection of P58IPK-silenced Nicotiana benthamiana and Arabidopsis knockouts leads to host death. This host cell death is associated with phosphorylation of the α subunit of eukaryotic translation initiation factor (eIF-2α). Loss of P58IPK leads to reduced virus titer, suggesting that wild-type P58IPK protein plays an important role in viral pathogenesis. Although our complementation results using mammalian P58IPK suggest conservation of the P58IPK pathway in plants and animals, its biological significance seems to be different in these two systems. In animals, P58IPK is recruited by the influenza virus to limit PKR-mediated innate antiviral response. In plants, P58IPK is required by viruses for virulence and therefore functions as a susceptibility factor.

|
Scooped by
Kamoun Lab @ TSL
June 7, 2012 7:55 AM
|
Plants lack the seemingly unlimited receptor diversity of a somatic adaptive immune system as found in vertebrates and rely on only a relatively small set of innate immune receptors to resist a myriad of pathogens. Here, we show that disease-resistant tomato plants use an efficient mechanism to leverage the limited nonself recognition capacity of their innate immune system. We found that the extracellular plant immune receptor protein Cf-2 of the red currant tomato (Solanum pimpinellifolium) has acquired dual resistance specificity by sensing perturbations in a common virulence target of two independently evolved effectors of a fungus and a nematode. The Cf-2 protein, originally identified as a monospecific immune receptor for the leaf mold fungus Cladosporium fulvum, also mediates disease resistance to the root parasitic nematode Globodera rostochiensis pathotype Ro1-Mierenbos. The Cf-2–mediated dual resistance is triggered by effector-induced perturbations of the apoplastic Rcr3pim protein of S. pimpinellifolium. Binding of the venom allergen-like effector protein Gr-VAP1 of G. rostochiensis to Rcr3pim perturbs the active site of this papain-like cysteine protease. In the absence of the Cf-2 receptor, Rcr3pim increases the susceptibility of tomato plants to G. rostochiensis, thus showing its role as a virulence target of these nematodes. Furthermore, both nematode infection and transient expression of Gr-VAP1 in tomato plants harboring Cf-2 and Rcr3pim trigger a defense-related programmed cell death in plant cells. Our data demonstrate that monitoring host proteins targeted by multiple pathogens broadens the spectrum of disease resistances mediated by single plant immune receptors.

|
Scooped by
Kamoun Lab @ TSL
June 5, 2012 9:25 AM
|
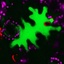
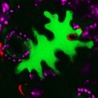
Effector assays: Green islands following agroinfiltration of tobacco (2012)
A warning about inadvertent effects of agroinfiltration in tobacco to add to the discussion "Some effector assays are prone to artefacts" (link below). Thanks to Jeff Ellis for contributing the image and the interpretation. Figure shows a tobacco leaf infiltrated with Agrobacterium tumefaciens GV3101 pMP90 resuspended at OD600=1 in 10 mM MgCl and containing various binary constructs. The plants were left in the glasshouse a long time after the experiment and had clearly run out of nutrients.The green island effect could be due to cytokinin made by A. tumefaciens. Nopaline strains do it in 2 ways, the tzs gene, which is acetosyringone inducible, and in the vir region of the Ti plasmid, and breakdown of tRNA. Transient assays of genes that are influenced by cytokinin would probably be affected and results could be confusing. More at http://storify.com/KamounLab/some-effector-assays-are-prone-to-artefacts
Earlier this month, the Gates Foundation awarded 107 researchers "Grand Challenges Exploration grants." 23 of those grants will address methods to protect crops from biotic stresses. The awards are well-deserved, innovative and exciting. Check them all out- here are a few: Taking Out the Bodyguards: A New Solution to Agricultural Disease - David Hughes, Penn State. Get rid of the ants (through RNAi) that protect the aphids that spread viruses and make honeydew substrates for pathogens. Field Selection of New Disease Resistance Alleles - Jonathan Jones, Sainsbury Lab, Norwich. Accelerate mutation rate in vivo of Resistance (R) genes to generate novel loci. Breeding the Epigenome in Crops - Sally Mackenzie, University of Nebraska. Disrupt the MSH1 gene to generate epigenetic variation, and screen for stress tolerance. Business Card-based Bacteriophage Therapy -Michael DuBow, Université Paris-Sud. Biodegradable cards impregnated with bateriophages that target plant pathogens. RNAi of the Rate Limiting Aflatoxin Biosynthesis Steps - Amos Alakonya, Jomo Kenyatta University of Agriculture and Technology, Nairobi. Maize producing RNAi to interefere with aflatoxin production. There are 18 more grants t0 amaze you, that include coatings to protect plants and seeds, rapid drying methods to prevent fungal growth post-harvest, lots of clever bio-control strategies, and even synthetic seeds. It's an inspiring set of people and projects - pass along to your students to motivate them!
Via Mary Williams

|
Scooped by
Kamoun Lab @ TSL
June 2, 2012 7:44 AM
|
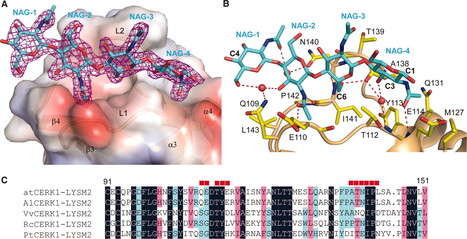
Pattern recognition receptors confer plant resistance to pathogen infection by recognizing the conserved pathogen-associated molecular patterns. The cell surface receptor chitin elicitor receptor kinase 1 of Arabidopsis (AtCERK1) directly binds chitin through its lysine motif (LysM)–containing ectodomain (AtCERK1-ECD) to activate immune responses. The crystal structure that we solved of an AtCERK1-ECD complexed with a chitin pentamer reveals that their interaction is primarily mediated by a LysM and three chitin residues. By acting as a bivalent ligand, a chitin octamer induces AtCERK1-ECD dimerization that is inhibited by shorter chitin oligomers. A mutation attenuating chitin-induced AtCERK1-ECD dimerization or formation of nonproductive AtCERK1 dimer by overexpression of AtCERK1-ECD compromises AtCERK1-mediated signaling in plant cells. Together, our data support the notion that chitin-induced AtCERK1 dimerization is critical for its activation.

|
Scooped by
Kamoun Lab @ TSL
May 29, 2012 5:01 PM
|
Post-conference thoughts on #OMGN12 Oomycete Molecular Genetics Network Annual Meeting, Nanjing, China
The conference was very much a success. There was notable progress relative to the 2011 Asilomar meeting. Newcomers to the field gave their first OMGN talks and the presentations by local scientists illustrated the wide-range of research taking place in China. I was sorry to miss the closing dinner but I suppose I saved myself the embarrassment of participating in the Karaoke. Below is an attempt to summarize and organize the somewhat random thoughts running through my head right now as I am sitting tight on the Shanghai to New York flight. Thanks to China Eastern Airlines for providing electrical outlets to all economy class passengers. The hot topic: epigenetics - The talks by Wenbo Ma and Mark Gijzen were the most thought provoking for me. Many questions are racing through my head as I am revisiting Wenbo’s talk. To what extent do oomycete effectors suppress RNA silencing? Did they evolve to directly target RNA silencing machinery? What about the nuclear-localized CRNs? Dinah Qutob and Mark Gijzen’s findings also raise many questions. What proportion of Phytophthora genes are targeted by silencing? How frequently do epialleles emerge? How frequently do they revert? What I found particularly exciting about both presentations are the questions they provoke. On the upswing: cell biology - Several presentations included a significant cell biology component but there is room for more. I expect cell biology to become increasingly integrated into oomycete research as we start considering the spatiotemporal aspects of the processes we study. As this happens we need to ensure that we use consistent standards and nomenclature. Not much new: genomics - I did not pick up any obvious new trends in genomics. The loss of heterozygosity (LOH) phenomenon described in a poster by Kurt Lamour et al. is probably the main novelty. There were some new genomes, e.g. Saprolegnia parasitica, Pythium insidiosum, and Pseudoperonospora cubensis. But some of these were described to some extent at previous OMGN conferences. I felt the topic was less dominant than in the past. The talk by the BGI representative was disappointing. What about apoplastic effectors? - How come so few are working on apoplastic effectors? None of the talks addressed these important players in oomycete-plant interactions. Mechanisms of RXLR effector translocation: the plot thickens - There is definitely more clarity than one year ago but many questions remain. The C-terminal binding of AVR3a/AVR1b to PI3Ps described by Yaeno et al. (2011) has been confirmed by Tyler at al. At least this one aspect does not seem to be controversial. But even if AVR3a/AVR1b C-terminal binding to PIPs mediates host cell translocation, it cannot be a general mechanism given that the C-termini of other RXLR effectors do not bind PIPs (I think but I am not sure that this point is accepted by several groups). Kasturi Haldar had some useful advice by reminding us that there are multiple routes to host cell entry in plasmodia. Is the RXLR leader generally involved in PIP binding? - A hotly debated issue remains the lack of reproducibility of the RXLR motif/domain binding to PIPs (Kale et al. 2010 vs. Yaeno et al. 2011 and unpublished reports). The first case of independent confirmation of this central finding of the Kale et al. paper is by Kasturi Haldar’s lab, which showed that the RXLR domain of P. infestans Nuk10 binds PI3Ps. However, the Haldar lab did not test the RXLR leader of AVR3a/AVR1b (pers. comm.), which are the leaders that could not be confirmed by Yaeno et al. and the van West lab (presented at OMGN11). Note also that the Nuk10 leader was not tested by Kale et al. Thus, in my view, the key finding of the Kale et al. paper - that the RXLR leader generally functions in PIP binding – still requires independent confirmation and would be nullified if indeed the AVR3a/AVR1b leader turns out to lack binding activity. What about Kale et al. experiments with full-length effectors? - The validated finding that the C-termini of AVR1b and related effectors bind PIPs compromises a number of experiments described in the Kale et al. paper. Assays in which RXLR mutations in full-length effectors abolish PI3P binding, as well as translocation into cells and roots, are inconsistent with the strong PIP binding of the C-termini. This casts an additional shadow on the quality of the Kale et al. experiments, which have already been plagued by the duplicated figures issue and lack of reproducibility of key assays with the fungal “RXLR-like” sequences. Other host-translocation leaders to the rescue? - Perhaps the breakthrough in understanding how oomycete effectors translocate inside host cells will come from other leaders than RXLR. Lobach, Wawra, van West et al. proposed that binding to tyrosine-O-sulphate on the host cell surface mediates entry of some Saprolegnia parasitica effectors. As far as I can tell this is not controversial although the finding has not been independently confirmed. Still, there is no work reported on other non-RXLR leaders, such as the Crinkler LxLFLAK and Albugo’s CHXC. Lets hope we’ll hear more about these in the future. Pre-secretion sorting? - I was impressed by the talk of Zhijian Zhao, which I thought was the most interesting of the several talks by members of the Nanjing Agricultural University oomycete group. Zhijian is investigating how oomycete effectors are secreted, focusing on the pathogen side. We’ll surely hear more on this topic in the future.

|
Rescooped by
Kamoun Lab @ TSL
from Plant Pathogenomics
May 28, 2012 2:16 PM
|
Meeting report of 2nd International Powdery Mildew Workshop and 3rd New Phytologist Workshop, in Zu¨ rich, Switzerland, February 2012.

|
Scooped by
Kamoun Lab @ TSL
May 26, 2012 7:53 PM
|
Day 2 Updates of #OMGN12 Oomycete Molecular Genetics Network Annual Meeting, Nanjing, China
|

|
Scooped by
Kamoun Lab @ TSL
June 14, 2012 11:34 AM
|

|
Scooped by
Kamoun Lab @ TSL
June 13, 2012 3:47 PM
|
Nature: Plant immunology: A life or death switch (2012)
Yay - the eagerly awaited top-10 list of plant pathogenic bacteria has been published! Can you guess which is number one? http://tinyurl.com/d4qjbmf This article joins the top-10 lists of fungal and viral pathogens, found here: http://onlinelibrary.wiley.com/journal/10.1111/%28ISSN%291364-3703/homepage/free_poster.htm These lists and the accompanying articles are great teaching resourses - what better way for students to learn about plant pathogens than to pick one from the list and study it? Students can write reviews or "press relases" on their pathogen, or make posters, or give five-slide talks. We've even heard of courses where students write songs or short plays to share what they've learned - engagement comes in many forms!
Via Mary Williams

|
Scooped by
Kamoun Lab @ TSL
June 11, 2012 6:14 AM
|
This outstanding book is an essential tool in helping the scientific community to identify smut fungi everywhere. It includes keys to the genera and species and a host plant–smut fungus list and compiles more than 3,500 micrographs and line drawings into a single sourcebook. This nearly 1,500-page treatise is authored by the worldwide authority on the subject, Dr. Kálmán Vánky, who has spent more than 50 years collecting and describing smut fungi species. The book provides complete and detailed presentations of species in 93 genera through descriptions and illustrations. The book updates our knowledge of valid scientific names and synonyms and provides taxonomic references and the host plant range of each species. Dr. Vánky’s passion for the topic shines through on every page through the excellent illustrations and detailed descriptions which complement each other to make identification easier. Having all content in a single volume allows the reader to flip back and forth between similar looking fungi for detailed comparison and analysis.

|
Scooped by
Kamoun Lab @ TSL
June 8, 2012 8:03 AM
|

|
Scooped by
Kamoun Lab @ TSL
June 10, 2012 10:02 AM
|
Video: Sophien Kamoun "The long reach of the effectors of plant associated organisms", 77th CSHL Symposium on Quantitative Biology: The Biology of Plants, June 1, 2012.

|
Scooped by
Kamoun Lab @ TSL
June 7, 2012 5:19 AM
|
Xinnian Dong chose to study plants in part because she’s uncomfortable with the alternative. She is fascinated by immunology, but the more obvious research models were not a good fit. “I feel squeamish working with animals, so I think plants are perfect,” she says. Thanks to that decision 20 years ago, today she is a pioneer in understanding how plant immunity works and spends her days probing the detailed mechanisms of plant–pathogen defense in her lab at Duke University. Postdoc positions available in Xinnian's lab. Please contact her directly.
Here are some resources for students considering a career in plant pathology. The first is the Careers page of the American Phytopathological Society (APS). There's a nice PDF booklet, as well as videos of plant pathologists describing their work. The British Society for Plant Pathology (BSPP) has a "week in the life of a plant pathologist" series here: http://www.bspp.org.uk/society/education.php
Via Mary Williams

|
Scooped by
Kamoun Lab @ TSL
June 3, 2012 9:29 AM
|
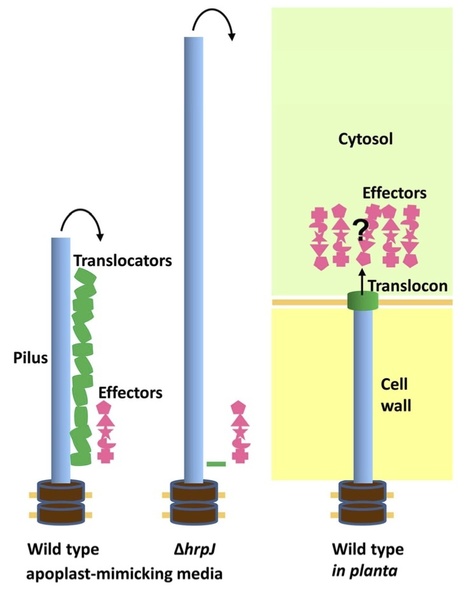
The assembly of type III secretion systems (T3SSs), which inject bacterial effector proteins into the cytosol of animal and plant hosts, is a highly regulated process. Animal pathogens use a length-control protein to produce T3SS needles of fixed length and then a second regulator, such as YopN in Yersinia spp, to mediate host contact-dependent effector delivery. For Pseudomonas syringae and other plant pathogens, regulation of the assembly process differs because the T3SS pilus must grow through variably thick plant cell walls before contacting the host plasma membrane. In this issue of Molecular Microbiology, Crabill et al. (2012) report evidence that the YopN homolog HrpJ is a multifunctional regulator of T3SS assembly in DC3000. A hrpJ mutant hyper-secretes pilus protein and no longer secretes four translocator proteins in culture, and it fails to inject effectors in planta. As with other proteins in this class, HrpJ is itself a T3SS substrate, but secretion-incompetent forms retain regulatory function. However, HrpJ is unusual in suppressing innate immune responses within host cells, as demonstrated with transgenic plants. The multiple capabilities of HrpJ appear to couple host contact sensing with pilus length control and translocator secretion while also contributing to immunity suppression early in the interaction.

|
Scooped by
Kamoun Lab @ TSL
May 31, 2012 7:34 AM
|
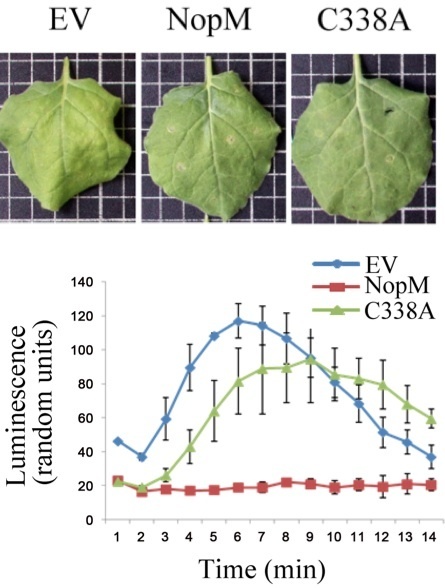
Type 3 effector proteins secreted via the bacterial type 3 secretion system (T3SS) are not only virulence factors of pathogenic bacteria, but also influence symbiotic interactions between nitrogen-fixing nodule bacteria (rhizobia) and leguminous host plants. In this study, we characterized NopM (nodulation outer protein M) of Rhizobium sp. strain NGR234, which shows sequence similarities with novel E3 ubiquitin ligase (NEL) domain effectors from the human pathogens Shigella flexneri and Salomonella enterica. NopM expressed in Escherichia coli, but not the non-functional mutant protein NopM-C338A, showed E3 ubiquitin ligase activity in vitro. In vivo, NopM, but not inactive NopM-C338A, promoted nodulation of the host plant Lablab purpureus by NGR234. When NopM was expressed in yeast, it inhibited mating pheromone signaling, a mitogen-activated protein (MAP) kinase pathway. When expressed in the plant Nicotiana benthamiana, NopM inhibited one part of the plant's defense response, as shown by a reduced production of reactive oxygen species (ROS) in response to the flagellin peptide flg22, whereas it stimulated another part, namely the induction of defense genes. In summary, our data indicate the potential for NopM as a functional NEL domain E3 ubiquitin ligase. Our findings that NopM dampened the flg22-induced ROS burst in N. benthamiana but promoted defense gene induction are consistent with the concept that pattern-triggered immunity is split in two separate signaling branches, one leading to ROS production and the other to defense gene induction.

|
Scooped by
Kamoun Lab @ TSL
May 28, 2012 2:38 PM
|
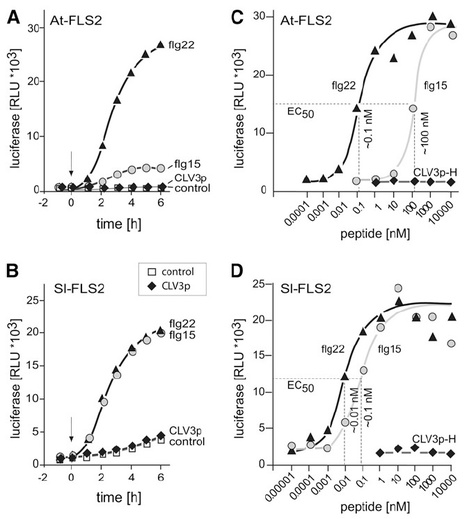
The flagellin receptor of Arabidopsis thaliana, At-FLAGELLIN SENSING2 (FLS2), has become a model for mechanistic and functional studies on plant immune receptors. Here, we started out with a comparison of At-FLS2 and the orthologous tomato (Solanum lycopersicum) receptor Sl-FLS2. Both receptors specifically responded to picomolar concentrations of the genuine flg22 ligand but proved insensitive to >106-fold higher concentrations of CLV3 peptides that have recently been reported as a second type of ligand for At-FLS2. At-FLS2 and Sl-FLS2 exhibit species-specific differences in the recognition of shortened or sequence-modified flg22 ligands. To map the sites responsible for these species-specific traits on the FLS2 receptors, we performed domain swaps, substituting subsets of the 28 leucine-rich repeats (LRRs) in At-FLS2 with the corresponding LRRs from Sl-FLS2. We found that the LRRs 7 to 10 of Sl-FLS2 determine the high affinity of Sl-FLS2 for the core part RINSAKDD of flg22. In addition, we discovered importance of the LRRs 19 to 24 for the responsiveness to C-terminally modified flagellin peptides. These results indicate that ligand perception in FLS2 is a complex molecular process that involves LRRs from both the outermost and innermost LRRs of the FLS2 ectodomain.

|
Scooped by
Kamoun Lab @ TSL
May 27, 2012 7:46 PM
|
Day 3 Updates of #OMGN12 Oomycete Molecular Genetics Network Annual Meeting, Nanjing, China
|

 Your new post is loading...
Your new post is loading...