Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
February 9, 5:23 AM
|
Workshop: Flow cytometry applied to microbiology (May 2026)

|
Scooped by
I2BC Paris-Saclay
January 28, 8:19 AM
|
Crosstalk between cohesins and axis proteins determines meiotic chromosome architecture in Sordaria macrospora
New discovery: a dynamic interplay between axis proteins and cohesins ensures chromosome stability during meiosis in Sordaria. Faithful chromosome segregation during meiosis requires the coordinated action of cohesin complexes and chromosome axis proteins. How these factors interact and communicate along chromosome axes, especially during meiotic prophase I, remains however, only partially understood. Researchers of the Genome Biology Department of the I2BC investigated the functional interplay between the cohesin components and regulators (Rad21, Rec8, Wapl, Sororin, Spo76/Pds5) and two meiosis-specific axis proteins Red1 and Hop1. Analysis of multiple combinations of their corresponding null mutants and of their genetic-epistasis interactions in the fungus Sordaria macrospora revealed a hierarchical regulatory network for their recruitment and releasing. Their work uncovers an unexpected role of axis proteins Red1 and Hop1, that together with Sororin, provide stage-specific protection of Spo76/Pds5 against Wapl-mediated release. Furthermore, we identify that Spo76/Pds5 is the main target of Wapl and acts as a central guardian of kleisin stability against Slx8/STUbL-dependent proteasomal degradation. Together, these findings show that a dynamic crosstalk between axis proteins and cohesins is crucial to preserve axis integrity and to ensure accurate meiotic progression. More information: https://doi.org/10.1371/journal.pgen.1012001 Contact: Stéphanie BOISNARD stephanie.boisnard@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 13, 3:30 PM
|
How virulence genes reorganize the Salmonella genome
Using functional genomics in sorted Salmonella populations and high-resolution microscopy, the researchers of the I2BC show that the expression of Pathogenicity Island 1 is associated with chromatin remodeling and with the repositioning of this region toward the nucleoid periphery. Chromatin provides a universal framework for organizing and regulating genomes across the three domains of life. In bacteria, it is composed of intrinsically supercoiled DNA associated with small DNA-binding proteins known as nucleoid-associated proteins (NAPs). Their binding can induce DNA bending, bridging, coating, and/or wrapping, giving rise to distinct modes of chromatin organization.
Bacterial chromatin can exist in a repressed state (silent chromatin) or in an actively transcribed state (active chromatin). Silent chromatin is largely associated with H-NS, a xenogeneic silencer that restricts the costly expression of genes acquired by horizontal transfer. In contrast, active chromatin is densely occupied by RNA polymerase and is characterized by different levels of DNA supercoiling. However, the changes in protein occupancy and chromatin organization that accompany transitions between these two states remain poorly understood.
Researchers of the Genome Biology Department of the I2BC in collaboration with the NGS and the Imaging facility of the I2BC and the Trinity College Dublin (Ireland), have unveiled the chromatin organization of horizontally acquired regions in Salmonella enterica serovar Typhimurium, which are essential for its pathogenicity. They show that expression of Pathogenicity Island 1 (SPI-1) is accompanied by local chromatin remodeling, marked by profound changes in three-dimensional organization and protein occupancy. This remodeling is also associated with the repositioning of SPI-1 toward the nucleoid periphery.
These findings provide new insights into the interplay between xenogeneic silencing, counter-silencing mechanisms, chromatin architecture, and the evolutionary integration of acquired DNA. They also reveal a finely tuned chromatin remodeling process that minimizes the cellular cost of activating pathogenicity islands, and they establish a direct link between the linear (1D) organization of the genome and its three-dimensional (3D) folding. More information: https://www.nature.com/articles/s41467-025-67746-w https://www.insb.cnrs.fr/fr/cnrsinfo/comment-les-genes-de-virulence-reorganisent-le-genome-de-salmonella-0 Contact: Vicky Lioy virginia.lioy@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 12, 7:46 AM
|
Cross-regulation of [2Fe–2S] cluster synthesis by ferredoxin-2 and frataxin.
Tight regulation of Fe-S clusters biosynthesis via a mutually antagonistic binding of frataxin and ferredoxin-2 to the assembly machinery, with several important implications for the Friedreich’s ataxia disease caused by frataxin deficiency. Iron-sulfur (Fe-S) clusters are essential metallocofactors that perform a multitude of biological functions. They are synthesized de novo by multi-proteins machienries and any defect in their synthesis leads to severe diseases such as Friedreich’s ataxia (FRDA), caused by defective expression of frataxin (FXN). Here, we uncover that efficient [2Fe-2S] cluster assembly requires a fine-tuned balanced ratio of FXN and Ferredoxin-2 (FDX2), an essential enzyme of the assembly process. [2Fe-2S] clusters are assembled on the scaffold protein ISCU2 with sulfur provided as a persulfide by NFS1, which is cleaved into sulfide by FDX2. FXN stimulates the whole process by accelerating persulfide transfer to ISCU2. Using an in vitro reconstituted human system, we show that any deviation from a close-to-equal amount of FXN or FDX2 downregulates Fe-S cluster synthesis. We performed a structure-function investigation, which revealed that this is due to competition between FXN and FDX2 for the same binding site on the NFS1-ISCU2 complex. We found that higher levels of FXN impair the persulfide-reductase activity of FDX2 and higher levels of FDX2 slow FXN-accelerated persulfide transfer to ISCU2. We also discovered that FDX2 directly hinders persulfide generation and transfer to ISCU2 by interacting with the persulfide-carrying mobile loop of NFS1. We further found that knocking-down FDX2 expression in a FRDA drosophila model, increases fly lifespan. Altogether, this work highlights a direct regulation of Fe-S cluster biosynthesis through antagonistic binding of FXN and FDX2 and suggests that decreasing FDX2 in the context of FXN deficiency in FRDA might constitute a novel therapeutic axis. More information: https://www.nature.com/articles/s41586-025-09822-1 Contact : Benoit D'Autréaux benoit.dautreaux@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 9, 5:05 AM
|
Gain and loss of gene function shaped the nickel hyperaccumulation trait in Noccaea caerulescens
Plants that hyperaccumulate nickel open the possibility to mine this metal from soils in an environmetally friendly manner. In this study, sequencing of a nickel hyperaccumulating plant together with genomic and transcriptomic comparisons reveal the molecular mechanisms underlying this extreme trait. Nickel hyperaccumulation is an extreme adaptation to ultramafic soils observed in more than 500 plant species. However, our understanding of the molecular mechanisms underlying the evolution of this trait remains limited. To shed light on these mechanisms, we have generated a high-quality genome assembly of the metal hyperaccumulator Noccaea caerulescens. We then used this genome as reference to conduct comparative intraspecific and interspecific transcriptomic analyses using various accessions of N. caerulescens and the non-accumulating relative Microthlaspi perfoliatum to identify genes associated with nickel hyperaccumulation.
Our results suggest a correlation between nickel hyperaccumulation and a decrease in the expression of genes involved in defense responses and the regulation of membrane trafficking. Surprisingly, these analyses did not reveal a significant enrichment of genes involved in the regulation of metal homeostasis. However, we found that the expression levels of selected metal transporter genes,
namely, NcHMA3, NcHMA4, and NcIREG2, are consistently elevated in N. caerulescens accessions hyperaccumulating nickel.
Furthermore, our analyses identified frameshift mutations in NcIRT1 associated with the loss of nickel hyperaccumulation in a few accessions. We further showed that the expression of a functional NcIRT1 in the roots of the La Calamine accession increases nickel accumulation in shoots. Our results demonstrate that NcIRT1 participates in nickel hyperaccumulation in N. caerulescens. They also
suggest that nickel hyperaccumulation is an ancient trait in N. caerulescens that has evolved from the high and constitutive expression of several metal transporters, including NcIREG2, and that the trait was subsequently lost in a few accessions due to mutations in NcIRT1. More information: https://doi.org/10.1093/plcell/koaf281 Contacts: Sylvain Merlot, sylvain.merlot@univ-tlse3.fr Sébastien Thomine, sebastien.thomine@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 10, 2025 4:16 AM
|
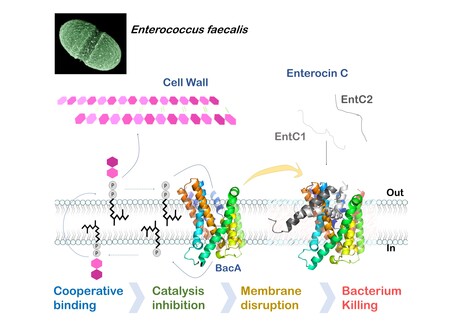
Mechanistic insights into Enterocin C bacteriocin targeting Enterococcus faecalis
Hijacking mechanism of membrane undecaprenyl phosphate recycling protein BacA in Enterococcus faecalis by a two-peptide bacteriocin revealed Antibiotic-resistance is a critical global health concern stressing the urgent need for new therapeutic strategies beyond conventional drugs. In this context, peptide-based bacteriocins constitute potential medical use antibacterial alternatives. In the present study, we have highlighted the molecular mechanism of a two-peptide bacteriocin, enterocin C, with potent activity against Enterococcus faecalis, a major opportunistic pathogen notorious for multidrug resistance. In collaboration with a group from INRAE MICALIS, the team has uncovered how enterocin C specifically exploits the membrane-embedded protein BacA as a cell surface receptor. BacA, widely conserved across bacteria, plays a central role in bacterial cell-wall biogenesis through its dual phosphatase and flippase activities, which are essential for recycling the lipid carrier undecaprenyl phosphate.
Using a combination of biochemical, biophysical, and microbiological approaches supported by AlphaFold structural modelling, we dissected the cooperative action of the two enterocin C peptides. Acting at nanomolar concentrations, peptide EntC1 inserts deeply into BacA’s outward-open catalytic pocket, blocking its enzymatic function and facilitating the subsequent binding of peptide EntC2. This dual docking event anchors the bacteriocin deep within the membrane’s hydrophobic core, ultimately triggering membrane disruption and bacterial cell death.
The findings reveal the molecular determinants of this precision targeting and represent the first detailed mechanistic description of a two-peptide bacteriocin’s mode of action. This work identifies BacA as a valuable target for bacteriocin-mediated killing and open avenues for the rational design of peptide-based antimicrobials for tailor-made antimicrobials to help combat antibiotic-resistant infections. More information : Thierry Touzé :thierry.touze@i2bc.paris-saclay.fr https://pubmed.ncbi.nlm.nih.gov/41232670/

|
Scooped by
I2BC Paris-Saclay
December 8, 2025 4:54 AM
|
Selective elimination of donor bacteria enables global profiling of plasmid gene expression during conjugation
A new ED-TA method which enabled genomics investigation of plasmid establishment during conjugative transfer was developed by I2BC researchers. They showed robust induction of a subset of plasmid genes at the early stages of conjugation through single-stranded DNA promoters. Bacterial conjugation is a principal mode of horizontal gene transfer which has important life science implications including bacterial genome evolution, dissemination of genetic traits and bioengineering applications. Notably, the spread of multidrug resistance via conjugative plasmids is one of the biggest concerns in global public health. Although conjugation has been studied since 1940s and the overall procedure is widely known and well documented, molecular details of reactions that establishes a transferred plasmid in the new host cell remain elusive. In addition, there are specific regulatory mechanisms for temporal expression of plasmid genes, that are also crucial for successful conjugation as they allow timely expression and function of plasmid-encoded arsenal against host defense mechanisms. Genomics-based studies of plasmid establishment were previously hampered by the nature of conjugation which takes place within a mixture of cell populations. Essentially, they require the separation of subpopulations before the DNA/RNA extraction to avoid contamination of indistinguishable plasmid DNA/RNA from the donor. Researchers of the Genome Biology Department of the I2BC, described the development of a new method, called ED-TA, which exploits a donor mutant hypersensitive to hypoosmotic shock. ED-TA allows unprecedently quick and efficient « Elimination of Donor population for Transconjugant Analysis». Using a clinically relevant model multidrug resistant conjugative plasmid, pESBL, they elucidated its transcription profile in successful and abortive conjugation. Researchers also showed single-stranded DNA promoters allow robust induction of a subset of genes at the early stages of conjugation. As the ED-TA method is straightforward and broadly applicable, further research taking advantage of the method will shed light on important molecular mechanisms of plasmid establishment after conjugation. More information: doi.org/10.1093/nar/gkaf1299 https://www.insb.cnrs.fr/fr/cnrsinfo/une-nouvelle-methode-pour-suivre-de-pres-la-transmission-des-genes-chez-les-bacteries Contact: Yoshiharu Yamaichi yoshiharu.yamaichi@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 1, 2025 4:49 AM
|
The Biophysics facility welcomes Viola Caroline D'mello
The Biophysics facility is pleased to welcome Viola Caroline D’mello, who joined in March 2025 as a CEA Ingénieure Chercheuse. After obtaining her Bachelor’s degree in Chemistry from Mumbai University and her Master’s in Analytical Chemistry from Mangalore University, she pursued her doctoral studies at the Tata Institute of Fundamental Research (TIFR) in Mumbai. Her thesis focused on the gas-phase study of hydrogen bonds in nitrogen-containing aromatic molecules using nanosecond UV and IR spectroscopy — hydrogen bonds similar to those found in DNA and RNA. In 2019, she joined LIDYL research group at Institute IRAMIS-CEA Saclay as a postdoctoral researcher, where she studied ion pairs and peptide folding in the gas phase. After a short period of working in industry in Bangalore, India, she returned to academia as a postdoctoral scholar in the Department of Physics at the University of Gothenburg, where she contributed to the commissioning of a high-resolution mass spectrometer coupled to a supersonic jet source. Since joining the Biophysics facility, she has taken charge of the electronic, Raman, and infrared spectroscopy facilities, overseeing user access while ensuring that experiments are designed and conducted under optimal conditions. She routinely supports and advises users on a wide range of techniques, including UV–Vis absorption, ultrafast transient absorption (from femtoseconds to milliseconds), FTIR, and Raman spectroscopy. Her broad expertise in photophysical methods, together with her reliability and open, collaborative attitude, makes her a central pillar of the Biophysics facility. More information : https://www.i2bc.paris-saclay.fr/biophysics/
Contact : biophysics@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
November 13, 2025 7:31 AM
|
The PIM facility welcomes Anaëlle and a new instrument: the Prometheus PANTA
A new piece of equipment was recently acquired by the Macromolecular Interactions Measurement facility (PIM) at the I2BC: the Prometheus PANTA, developed by Nanotemper Technologies. The Prometheus PANTA combines several detection methods to simultaneously measure thermal stability and aggregation tendency. Its technology is mainly based on nanoDSF (Differential Scanning Fluorimetry), which detects changes in protein conformation as a function of heating temperature, without the need for fluorescent labeling. The intrinsic fluorescence of aromatic amino acids (tryptophan and tyrosine) serves as a probe to track thermal transitions and determine denaturation points (Tm), which are direct indicators of protein stability. In addition, the system incorporates Dynamic Light Scattering (DLS) to measure hydrodynamic radius and detect oligomer formation, as well as a Back Reflection measurement that provides information on solution turbidity and protein aggregation. These combined measurements provide a global view of protein behavior under different experimental conditions, with precise temperature control (from 15 to 110°C) and excellent reproducibility. Each analysis requires only a few microliters of sample (10µL per capillary). The Prometheus PANTA is therefore a versatile tool for studying biomolecules. It allows you to: -
Evaluate the thermal stability of proteins -
Study interactions between proteins and a ligand -
Optimize buffer formulation, purification, and storage conditions -
Monitor the formation of aggregates or condensates under varying temperature or composition conditions. Its sensitivity, speed of analysis, and ability to measure multiple samples, up to 48 capillaries in parallel, make it a tool suitable for both fundamental research and biotechnology or structural biology projects. The arrival of Prometheus PANTA is accompanied by that of Anaëlle KALUBI KABAMBI, an assistant engineer recently recruited to the PIM facility. She will be responsible for implementing and optimizing analysis protocols, as well as assisting users in sample preparation and data interpretation. Her expertise in biochemistry will strengthen the facility’s capabilities, helping to provide researchers with comprehensive support for characterizing the interactions and structural properties of biomolecules Complementing the other instruments available at PIM (DSC, ITC, BLI, FIDA, and MST), the Prometheus PANTA enhances the facility’s ability to offer a complete range of protein characterization techniques. More information: https://www.i2bc.paris-saclay.fr/structural-biology/pim/ Contacts: anaelle.kalubi-kabambi@i2bc.paris-saclay.fr ; stephanie.marsin@i2bc.paris-saclay.fr ; magali.aumont-nicaise@i2bc.paris-saclay.fr ; magali.noiray@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 15, 2025 9:07 AM
|
Moss BRCA2 lacking the canonical DNA-binding domain promotes homologous recombination and binds to DNA
Despite having low homology with BRCA2 proteins from other organisms and no folded domain, the newly discovered BRCA2 protein from Physcomitrium patens (the green yeast) promotes homologous recombination by binding to recombinases and DNA. BRCA2 is crucial for mediating homology-directed DNA repair (HDR) through its binding to single-stranded DNA (ssDNA) and the recombinases RAD51 and DMC1. Most BRCA2 orthologs have a canonical DNA-binding domain (DBD) with the exception of Drosophila melanogaster. It remains unclear whether such a noncanonical BRCA2 variant without DBD possesses a DNA-binding activity. Here, we identify a new noncanonical BRCA2 in the model plant Physcomitrium patens (PpBRCA2). We establish that PpBRCA2 is essential for genome integrity maintenance, somatic DNA double-strand break (DSB) repair, HDR-mediated gene targeting, and RAD51 foci recruitment at DNA break sites. PpBRCA2 is also critical for DSB repair during meiosis. Interestingly, PpBRCA2 interacts strongly with RAD51 but weakly with DMC1, suggesting a distinct meiotic function compared to other BRCA2 homologs. Despite lacking the canonical DBD, PpBRCA2 binds ssDNA through its disordered N-terminal region and efficiently promotes HDR. Our work highlights that the ssDNA binding capacity of BRCA2 homologs is conserved regardless of the presence of a canonical DBD and provides a deeper understanding of BRCA2’s functional diversity across species. More information: https://pmc.ncbi.nlm.nih.gov/articles/PMC12412785/ Contact: Sophie Zinn sophie.zinn@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 5, 2025 3:50 AM
|
Restriction of Ku translocation protects telomere ends
How Ku inward translocation can be restricted to protect telomere ends from NHEJ without altering other important functions. Safeguarding chromosome ends against fusions via nonhomologous end joining (NHEJ) is essential for genome integrity. Paradoxically, the conserved NHEJ core factor Ku binds telomere ends. How it is prevented from promoting NHEJ remains unclear, as does the mechanism that allows Ku to coexist with telomere-protective DNA binding proteins, Rap1 in Saccharomyces cerevisiae. Here, we find that Rap1 directly inhibits Ku’s NHEJ function at telomeres. A single Rap1 molecule near a double-stand break suppresses NHEJ without displacing Ku in cells. Furthermore, Rap1 and Ku form a complex on short DNA duplexes in vitro. Cryo-EM shows Rap1 blocks Ku’s inward translocation on DNA – an essential step for NHEJ at DSBs. Nanopore sequencing of telomere fusions confirms this mechanism protects native telomere ends. These find- ings uncover a telomere protection mechanism where Rap1 restricts Ku’s inward translocation. This switches Ku from a repair-promoting to a protective role preventing NHEJ at telomeres. More information : https://doi.org/10.1038/s41467-025-61864-1 Contact : Philippe Cuniasse philippe.cuniasse@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
August 20, 2025 3:34 AM
|
Confocal microscopy workshop 2025

|
Scooped by
I2BC Paris-Saclay
June 30, 2025 5:14 AM
|
Training session on TEM in cell biology - 14-17 October 2025
|

|
Scooped by
I2BC Paris-Saclay
February 9, 3:57 AM
|
Cytometry workshop (March 2026)

|
Scooped by
I2BC Paris-Saclay
January 13, 3:52 PM
|
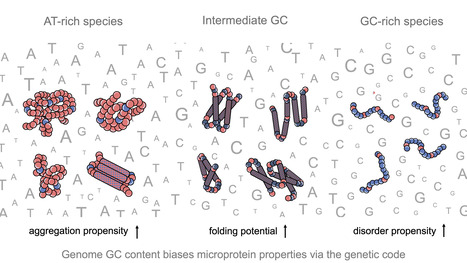
Impact of GC content on de novo gene birth
Noncoding DNA can generate microproteins, some of which evolve into new genes. We show that de novo genes preferentially originate from GC-rich, foldable sequences, revealing how base composition channels the birth of new proteins. Noncoding regions of eukaryotic genomes are widely transcribed and constitute a major source of novel microproteins, some of which eventually become fixed as de novo genes - a process known as de novo gene birth that plays a significant role in species adaptation. However, the structural properties of these nascent proteins and the factors governing their evolutionary fate remain poorly understood. In particular, the role of genome nucleotide composition (GC content) in shaping their biophysical properties has remained unclear. In this study, researchers of the I2BC, analyzed the foldability and sequence properties of millions of putative microproteins encoded by intergenic open reading frames (ORFs) from 3,379 eukaryotic species spanning a broad range of GC contents (18–79%). Results show that GC content strongly influences amino-acid composition and structural tendencies, suggesting distinct cellular impacts if non-genic regions are pervasively expressed. AT-rich species predominantly encode ORFs biased toward hydrophobic, aggregation-prone sequences, whereas GC-rich species tend to encode more hydrophilic, disorder-prone ORFs. ORFs from genomes with intermediate GC content display a more balanced composition and higher folding potential, with many expected to adopt proto-folds. To assess how these properties relate to gene emergence,the authors traced the evolutionary history of several hundred de novo proteins across 22 species using phylostratigraphy, targeted de novo gene searches, and ancestral sequence reconstruction. Researchers find that de novo genes preferentially originate from GC-rich ORFs with intrinsic folding potential. Together, our results reveal that the interplay between GC content and foldability - rooted in the structure of the genetic code - shapes the emergence of novel genes. More information: https://www.nature.com/articles/s41467-025-68022-7 Contact: Anne Lopes anne.lopes@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 13, 5:33 AM
|
Flexizyme-Based Strategy for the Synthesis of Stable, Non-Isomerizable Amide-Linked 2’-Aminoacyl-tRNAs and their Shortened Analogs
We developed a flexizyme-based semi-synthetic strategy that provides access to stable 2′- and 3′-amide AA-tRNA analogs, offering robust tools for structural biology and for probing regiospecificity in AA-tRNA-dependent enzymes. The study of the regiospecificity of aminoacyl-tRNA (AA-tRNA)-dependent enzymes and their structural characterization with AA-tRNAs are limited by rapid hydrolysis of the ester bond linking amino acid to tRNA. To overcome this limitation, stable AA-tRNA analogs bearing hydrolysis-resistant linkages, such as amide bonds or ester bioisosteres, have been developed. These analogs are valuable tools for investigating interactions between AA-tRNAs and various enzymes or ribonucleoproteins, including elongation factors, ribosomes, Fem-family transferases, and cyclodipeptide synthases. However, their synthesis remains technically challenging. Recently, flexizymes—engineered ribozymes capable of aminoacylating tRNAs with diverse amino acids or analogs—have enabled the synthesis of 3′-amide-linked AA-NH-tRNAs. Due to their inherent specificity for 3′-OH acylation, flexizymes have not been used to generate 2′-amide-linked analogs, and such regioisomers have remained unexplored. In this study, we demonstrate that while flexizymes cannot directly aminoacylate the 2′ position, they can nevertheless mediate the synthesis of 2′-aminoacyl-NH-tRNAs via a two-step regioisomerization mechanism with excellent yields. This finding provides new insights into the binding mode of AA-tRNAs to flexizymes and expands the chemical space of stable AA-tRNA analogs. Access to both 3′- and 2′-amide regioisomers will enable more precise studies of AA-tRNA recognition and catalysis by various AA-tRNA-dependent systems. More information : https://doi.org/10.1002/chem.202503506 Contact person: Matthieu Fonvielle matthieu.fonvielle@i2bc.paris-saclay.fr https://www.i2bc.paris-saclay.fr/enzymology-and-non-ribosomal-peptide-biosynthesis/

|
Scooped by
I2BC Paris-Saclay
January 9, 5:24 AM
|
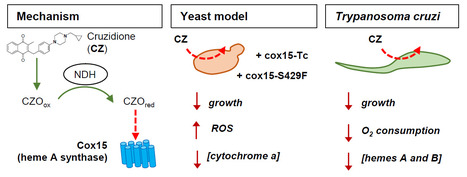
The heme A synthase Cox15, as a target of redox-active 3-benzylmenadiones with antiparasitic activity
We identified a new target for antiparasitic compounds and it is in the mitochondria. Chagas disease, caused by Trypanosoma cruzi, is a neglected parasitic infection. The very limited arsenal of anti-T. cruzi treatments calls for the development of new drugs. Recently, a library of 3-benzylmenadione derivatives was synthesized with cruzidione being the most efficient and specific compound against the parasite. To decipher its mode of action, we used the yeast Saccharomyces cerevisiae as model. Evidence pinpointed at the heme A synthase Cox15 as a primary target of cruzidione: 1) a mutation in Cox15 (i.e., S429F) renders the yeast cells highly sensitive to the drug, 2) treatment with cruzidione led to the loss of cytochrome c oxidase, an enzyme that relies on heme A as an essential cofactor and 3) replacement of the yeast Cox15 by T. cruzi enzyme resulted in a high sensitivity to cruzidione. We then investigated the effect of cruzidione in T. cruzi and observed a significant reduction of heme contents, most likely involving the inhibition of the heme A synthase. This, in turn, led to a decrease in O2 consumption by the parasite. Finally, using the yeast model, we showed that, similarly to what we previously found for the antimalarial benzylmenadione plasmodione, NADH-dehydrogenase plays a key role in cruzidione bioactivation. We proposed that the reduced benzoylmenadione metabolites produced by the reaction with NADH-dehydrogenase, act as Cox15 inhibitors. This study, through the identification of the mode of action of cruzidione, highlighted Cox15 as a novel target for antiparasitic drugs. More information: https://journals.asm.org/doi/10.1128/aac.01161-25 Contact: Brigitte Meunier, brigitte.meunier@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 6, 4:23 PM
|
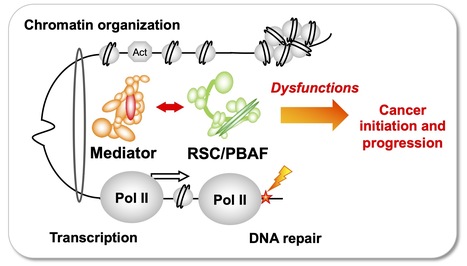
Team of J. Soutourina received the label from Ligue Nationale contre le cancer
Understanding how the essential complexes —the coregulator Mediator and the chromatin remodeling complex RSC/PBAF— cooperate in the nucleus In eukaryotes, transcription and DNA repair occur in the crowded context of chromatin. Dysfunctions of these processes can lead to cancers. Mediator is an essential and conserved multisubunit coactivator complex, mutated in many cancers. However, it remains largely unknown how Mediator and chromatin regulators coordinate their functions. A recent publication of the team suggests the novel hypothesis that Mediator acts in conjunction with the chromatin remodeling complex RSC (Remodels the Structure of Chromatin) of SWI/SNF family, homologous to PBAF (Polybromo-associated BAF) in human, representing the most frequently mutated complexes in cancers. Building on this recent publication, a new project has been supported by Ligue Nationale contre le cancer. Using the yeast model, with a perspective to extend the study to human cells, the team intends to decipher the molecular mechanisms involved in functional cooperation between these essential coregulator complexes in transcription regulation, DNA repair and chromatin organization relevant for cancer biology. Contact: Julie Soutourina julie.soutourina@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 8, 2025 5:52 AM
|
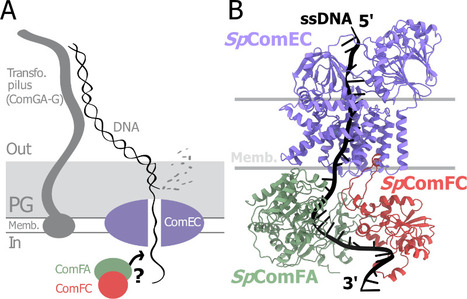
A tripartite protein complex promotes DNA transport during natural transformation in Firmicutes.
From structural modeling to natural transformation in bacteria: identification of a key protein complex for DNA transport across the membrane of Firmicutes. Transformation is a key mechanism of horizontal gene transfer, central to bacterial adaptation. This evolutionarily conserved process allows bacteria to integrate exogenous genetic material into their genome, thereby facilitating, for example, the spread of antibiotic resistance.
In a study published in the journal PNAS, scientists from I2BC (CEA/CNRS/UPSaclay, Gif-sur-Yvette) and DRCM/IBFJ (CEA/UPSaclay/UP Cité, Fontenay-aux-Roses), in collaboration with a laboratory at CBI (CNRS/University of Toulouse), identified a protein complex involved in the transport of single-stranded DNA across the membrane during transformation. Using AlphaFold, they structurally modeled this three-protein complex, which is highly conserved in the phylum Firmicutes, in interaction with single-stranded DNA. The structural model allowed them to identify a possible path for DNA through a conserved channel in one of the three proteins, a transmembrane protein, and then along a groove formed by the other two proteins. This model was validated by a robust experimental strategy in the bacterium Streptococcus pneumoniae, by measuring the impact on transformation efficiency of disruptive mutations in several protein–protein and protein–DNA interfaces. The transmembrane channel was found to be conserved in a structural model in Helicobacter pylori, and its importance for transformation was also experimentally confirmed in this bacterium. This study sheds light on the molecular mechanisms of bacterial transformation and demonstrates the power of macromolecular structure prediction to generate molecular hypotheses and guide functional experiments. More information: https://doi.org/10.1073/pnas.2511180122 Contact : Jessica Andreani jessica.andreani@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 5, 2025 9:28 AM
|
Structural determinants of SlpA-mediated phage recognition in Clostridioides difficile
Study published in PLoS Pathogens of molecular features of phage receptor SlpA protein demonstrated the complexity of the interactions between human enteropathogen C. difficile and its phages The emergence of Clostridioides difficile as a leading cause of healthcare-associated nosocomial intestinal infections calls for novel therapeutic strategies, particularly in the context of antibiotic resistance and recurrent disease. Phage therapy is a promising approach, but its clinical application against C. difficile remains hindered by our lack of understanding of the factors driving host specificity. Indeed, it is crucial to understand how phages specifically interact with their host to be able to select the best candidates for cocktail preparation. The main surface layer protein SlpA is a key phage receptor, but the molecular details governing phage-receptor interactions remain unclear. By dissecting the structural features of SlpA required for phage infection through engineered SlpA isoforms and domain modifications, in collaboration with our Canadian collegues from Sherbrooke University, we reveal how specific regions of SlpA mediate phage adsorption and infection. These new insights into phage–receptor interactions will be instrumental in guiding the future engineering of broad-host-range therapeutic phages. More information : https://doi. org/10.1371/journal.ppat.1013724 Contact : Olga Soutourina olga.soutourina@i2bc.paris-saclay.fr https://www.i2bc.paris-saclay.fr/regulatory-rnas-in-clostridia/

|
Scooped by
I2BC Paris-Saclay
November 20, 2025 6:14 AM
|
Strict gut symbiont specificity in Coreoidea insects governed by interspecies competition
Insects choose their bacterial gut symbionts by creating a multifactorial selective environment that promotes competition among symbiont candidates and colonization by the single, best-adapted strain

|
Scooped by
I2BC Paris-Saclay
November 9, 2025 4:21 AM
|
Three stop codons to get over to flourish
Funded by the European Research Council (ERC synergy 2025), the project “3Stops2Go” leaps over the “three red lights” of premature stop codons to re-express critical protein and correct genetic diseases. The project, recently funded by an ERC Synergy Grant 2025, aims to target premature termination codon (PTC)mutations, which prematurely halt protein translation and are involved in about 11% of human genetic diseases.
By combining expertise in protist biology, RNA biology, and gene therapy, the consortium of four researchers (Leoš Valášek (Institute of Microbiology, Czech Academy of Sciences, Czech Republic), Julius Lukeš (Biology Centre, Czech Academy of Sciences, Czech Republic), Olivier Namy (Université Paris-Saclay, CEA, CNRS, France / I2BC, France), and Mark Osborn (University of Minnesota, USA)) aims to harness natural stop-codon bypass mechanisms to develop therapeutic tools (engineered tRNAs and readthrough inducers), test them in patient-derived cells and animal models, and ultimately pave the way toward clinical applications. More information: https://erc.europa.eu/news-events/news/synergy-grants-2025-examples-projects Contact: Olivier Namy olivier.namy@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 6, 2025 3:37 PM
|
New team in the Department of Genome Biology
Cécile Courret, CNRS Researcher and recipient of an ATIP-Avenir grant, has joined the Department of Genome Biology to establish her team ‘Intragenomic Conflict and Evolution’. Cécile's research focus on genetic conflicts and their impact on genome evolution. The basic principles of Mendelian inheritance state that in heterozygous individuals, two alleles have equal chances of being transmitted to the next generation. However, some genetic elements do not follow these rules. These so-called selfish genetic elements, such as transposons or meiotic drivers, bias inheritance in their own favor, often at the expense of the organism. Because they disrupt essential processes like meiosis, heterochromatin regulation, and cell division, selfish elements create a persistent conflict with the host genome. This conflict triggers an evolutionary “arms race,” in which genomes evolve defense mechanisms to counterbalance the harmful effects of these elements. Far from being rare exceptions, such conflicts are now recognized as a major force shaping genome structure and function. My research relies on the Drosophila model and combines genomic, molecular, and cytological approaches to investigate both how selfish elements perturb fundamental biological processes and how organisms respond to mitigate these disruptions. By studying systems where these conflicts are still active in natural populations, as well as the genomic signatures left by past events, Cécile's team aims to better understand how conflicts drive evolutionary innovation and shed light on the molecular basis of essential biological mechanisms. Contact: Cécile Courret, cecile.courret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 10, 2025 6:12 AM
|
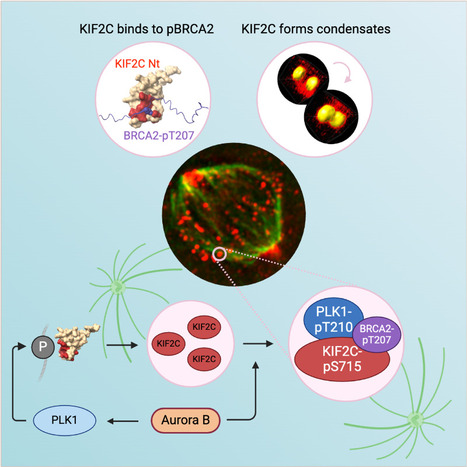
KIF2C condensation concentrates PLK1 and phosphorylated BRCA2 on kinetochore microtubules in mitosis
The microtubule depolymerase KIF2C forms membrane-less organelles at the kinetochore through its N-terminal phospho-peptide binding domain that interacts with BRCA2 and other phosphorylated targets in mitosis. During mitosis, the microtubule depolymerase KIF2C, the tumor suppressor BRCA2, and the kinase PLK1 contribute to the control of kinetochore-microtubule attachments. Both KIF2C and BRCA2 are phosphorylated by PLK1, and BRCA2 phosphorylated at T207 (BRCA2-pT207) serves as a docking site for PLK1. Reducing this interaction results in unstable microtubule-kinetochore attachments. Here we identified that KIF2C also directly interacts with BRCA2-pT207. Indeed, the N-terminal domain of KIF2C adopts a Tudor/PWWP/MBT fold that unexpectedly binds to phosphorylated motifs. Using an optogenetic platform, we found that KIF2C forms membrane-less organelles that assemble through interactions mediated by this phospho-binding domain. KIF2C condensation does not depend on BRCA2-pT207 but requires active Aurora B and PLK1 kinases. Moreover, it concentrates PLK1 and BRCA2-pT207 in an Aurora B-dependent manner. Finally, KIF2C depolymerase activity promotes the formation of KIF2C condensates, but strikingly, KIF2C condensates exclude tubulin: they are located on microtubules, especially at their extremities. Altogether, our results suggest that, during the attachment of kinetochores to microtubules, the assembly of KIF2C condensates amplifies PLK1 and KIF2C catalytic activities and spatially concentrates BRCA2-pT207 at the extremities of microtubules. We propose that this novel and highly regulated mechanism contributes to the control of microtubule-kinetochore attachments, chromosome alignment, and stability. More Information : https://academic.oup.com/nar/article/53/11/gkaf476/8160319 Contact: Sophie Zinn sophie.zinn@i2bc.paris-saclay.fr
|






 Your new post is loading...
Your new post is loading...


![Cross-regulation of [2Fe–2S] cluster synthesis by ferredoxin-2 and frataxin. | I2BC Paris-Saclay | Scoop.it](https://img.scoop.it/Ve3yN6adKt_5lzSWPuYkATl72eJkfbmt4t8yenImKBVvK0kTmF0xjctABnaLJIm9)