 Your new post is loading...

|
Scooped by
?
Today, 3:53 PM
|
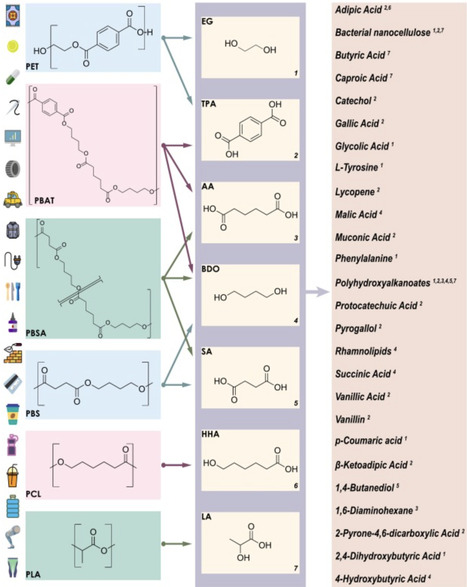
Since their large-scale adoption in the early 20th century, plastics have become indispensable to modern life. However, inadequate disposal and recycling methods have led to severe environmental consequences. While traditional end-of-life plastics management had predominantly relied on landfilling, a paradigm shift towards recycling and valorization emerged in the 1970s, leading to the development of various, mostly mechanochemical, recycling strategies, together with the more recent approach of biological depolymerization and upcycling. Plastic upcycling, which converts plastic waste into higher-value products, is gaining attention as a sustainable strategy to reduce environmental impact and reliance on virgin materials. Microbial plastic upcycling relies on efficient depolymerization methods to generate monomeric substrates, which are subsequently metabolized by native or engineered microbial systems yielding valuable bioproducts. This review focuses on the second phase of microbial polyester upcycling, examining the intracellular metabolic pathways that enable the assimilation and bioconversion of polyester-derived monomers into industrially relevant compounds. Both biodegradable and non-biodegradable polyesters with commercial significance are considered, with emphasis on pure monomeric feedstocks to elucidate intracellular carbon assimilation pathways. Understanding these metabolic processes provides a foundation for future metabolic engineering efforts, aiming to optimize microbial systems for efficient bioconversion of mixed plastic hydrolysates into valuable bioproducts.

|
Scooped by
?
Today, 3:49 PM
|
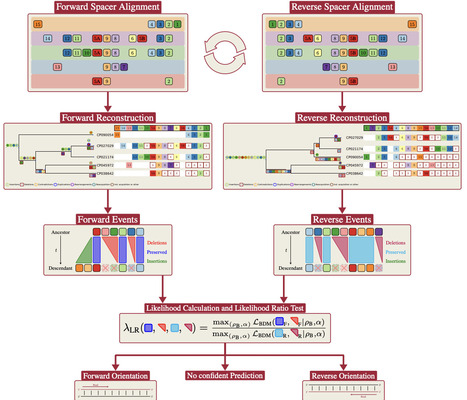
CRISPR-Cas is a defense system of bacteria and archaea against phages. Parts of the foreign DNA, called spacers, are incorporated into the CRISPR array which constitutes the immune memory. The orientation of CRISPR arrays is crucial for analyzing and understanding the functionality of CRISPR systems and their targets. Several methods have been developed to identify the orientation of a CRISPR array. To predict the orientation, different methods use different features such as the repeat sequences between the spacers, the location of the leader sequence, the Cas genes, or PAMs. However, those features are often not sufficient to predict the orientation with certainty, or different methods disagree. Remarkably, almost all CRISPR systems have been found to insert spacers in a polarized manner at the leader end of the array. We introduce CRISPR-evOr, a method that leverages the resulting patterns to predict the acquisition orientation for (a group of) CRISPR arrays by reconstructing and comparing the likelihood of their evolutionary history with respect to both possible acquisition orientations. The new method is independent of Cas type, leader existence and location, and transcription orientation. CRISPR-evOr is thus particularly useful for arrays that other CRISPR orientation tools cannot predict confidently and to verify or resolve conflicting predictions from existing tools. CRISPR-evOr currently confidently predicts the orientation of 28.3% of the arrays in the considered subset of CRISPRCasdb, which other tools like CRISPRDirection and CRISPRstrand cannot reliably orient. As our tool leverages evolutionary information we expect this percentage to grow in the future when more closely related arrays will be available. Additionally, CRISPR-evOr provides confident decisions for rare subtypes of CRISPR arrays, where knowledge about repeats and leaders and their orientation is limited.

|
Scooped by
?
Today, 3:38 PM
|
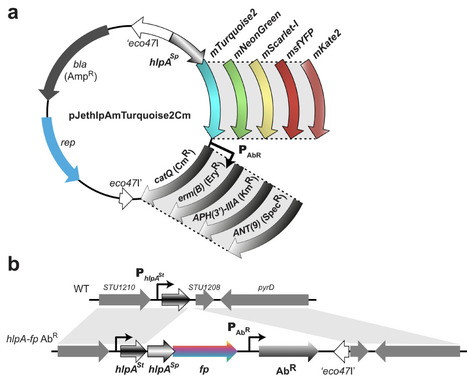
Fluorescent labeling is a powerful tool in microbiology allowing live cell imaging and providing insights into dynamic cellular processes, quantification of gene expression and protein subcellular localization. Although multicolor imaging is widely used in Streptococcus pneumoniae and S. mutans, variants other than the green fluorescent protein (GFP) have rarely been applied in other streptococcal species. To address this gap in the streptococcal molecular toolbox, we benchmarked five different fluorescent proteins. The various fluorescent proteins were fused to the C-terminus of S. pneumoniae HlpA, a small non-specific DNA binding histone-like protein. These reporters, combined with four different antibiotic resistance genes, were engineered with various expression systems (inducible or constitutive) to form versatile cassettes. We provide methods to transfer these cassettes to different streptococcal species including S. salivarius and S. thermophilus. As a proof of concept, we generated a triple labeled S. salivarius strain in which HlpA, FtsZ and DivIVA were fused to three spectrally-distinct compatible fluorescent proteins. Multiple fluorescent labeling has broad applications for deciphering a wide range of scientific problems, from cellular processes to infectious disease mechanisms. The availability of these cassettes should allow for a wider use of single-cell labeling strategies in the Streptococcus clade and other closely related bacteria.

|
Scooped by
?
Today, 3:26 PM
|
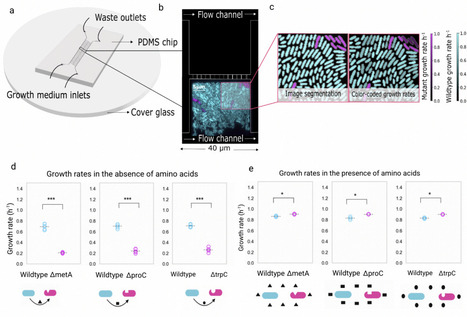
Most microbes grow in spatially structured, genetically diverse communities where they interact metabolically with neighboring cells. In such settings, evolutionary processes can result in the loss of biosynthetic pathways, leading to dependencies among strains. The Black Queen Hypothesis suggests that these gene losses reduce metabolic burden, potentially conferring a fitness advantage. However, how these dependencies evolve at the level of individual cells remains poorly understood. Here, we introduce an experimental system that enables direct observation of the first steps in the evolution of metabolic dependencies at single-cell resolution. Using a microfluidic setup combined with live-cell microscopy, we tracked the fate of individual amino acid auxotrophic mutant cells - deficient in methionine, tryptophan, or proline biosynthesis - within spatially structured Escherichia coli wildtype populations. When amino acids were externally supplied, auxotrophs grew faster than wildtype cells, Without external supply of amino acids, they grew significantly slower. When auxotrophs formed cell clusters, local depletion of amino acids further diminished their growth rates as well as the growth of neighbouring wildtype cells. Our mathematical model indicates that auxotroph growth is strongly constrained by the low rate of amino acid leakage from wildtype neighbors, conferring an advantage only once leakage surpasses a critical threshold. Overall, our findings show that the benefits of gene loss are highly context-dependent. Through single-cell experiments and modeling, we shed light on the early evolution of metabolic dependency and the trade-offs between reducing metabolic costs and maintaining metabolic autonomy in microbial communities.

|
Scooped by
?
Today, 3:16 PM
|
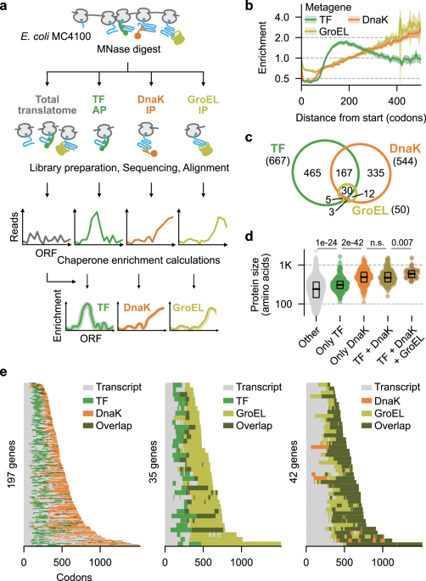
Chaperones are essential to the co-translational folding of most proteins. However, the principles of co-translational chaperone interaction throughout the proteome are poorly understood, as current methods are restricted to few substrates and cannot capture nascent protein folding or chaperone binding sites, precluding a comprehensive understanding of productive and erroneous protein biosynthesis. Here, by integrating genome-wide selective ribosome profiling, single-molecule tools, and computational predictions using AlphaFold we show that the binding of the main E. coli chaperones involved in co-translational folding, Trigger Factor (TF) and DnaK correlates with “unsatisfied residues” exposed on nascent partial folds – residues that have begun to form tertiary structure but cannot yet form all native contacts due to ongoing translation. This general principle allows us to predict their co-translational binding across the proteome based on sequence only, which we verify experimentally. The results show that TF and DnaK stably bind partially folded rather than unfolded conformers. They also indicate a synergistic action of TF guiding intra-domain folding and DnaK preventing premature inter-domain contacts, and reveal robustness in the larger chaperone network (TF, DnaK, GroEL). Given the complexity of translation, folding, and chaperone functions, our predictions based on general chaperone binding rules indicate an unexpected underlying simplicity. This study integrates ribosome profiling, single molecule methods and computational predictions to reveal that molecular chaperones bind ‘unsatisfied’ residues exposed on partial nascent folds, rationalizing how their activities generate a robust co-translational protein folding network.

|
Scooped by
?
Today, 2:50 PM
|
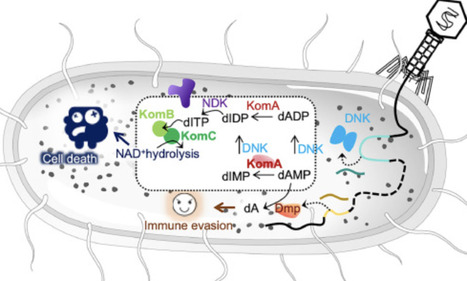
Bacteria employ diverse immune systems, such as CRISPR-Cas, to fend off phage infections. A recent study uncovered the unprecedented mechanistic features of the Kongming bacterial defense system, which uniquely exploits phage-derived enzymes to synthesize deoxyinosine triphosphate (dITP), thereby triggering host immunity through NAD+ depletion. In response, some phages have evolved countermeasures to disrupt dITP synthesis, highlighting the ongoing evolutionary arms race between hosts and pathogens. This discovery not only deepens our understanding of bacterial defense strategies but also paves the way for new insights in biomedical research and synthetic biology.

|
Scooped by
?
Today, 2:31 PM
|
Pesticides can modulate gut microbiota composition, but their specific effects on it remain largely elusive. In our study, we show that pesticides inhibit or promote the growth of various gut microbial species and can be accumulated to prolong their presence in the host. Pesticide exposure also induces significant alterations in gut bacterial metabolism, as reflected by changes in hundreds of metabolites. We generate a pesticide-gut microbiota-metabolite network that not only reveals pesticide-sensitive gut bacteria species but also reports specific metabolic changes in 306 pesticide-gut microbiota pairs. Using an in vivo mouse model, we further demonstrate the interactions of a representative pesticide-gut microbiota pair and verify the inflammation-inducing effects of pesticide exposure on the host, mediated by microbially dysregulated lipid metabolism. Taken together, our findings generate a comprehensive atlas of pesticide-gut microbiota-metabolite interactions atlas and shed light on the molecular mechanisms by which pesticides affect host health via gut microbiota-modulated metabolism. Pesticide exposure induces significant alterations in gut bacterial metabolism. Here, the authors perform a multi-omics mapping of pesticide-gut microbiota-metabolite interactions, identifying pesticide-sensitive gut bacteria species and specific metabolic changes in 306 pesticide-gut microbiota pairs.

|
Scooped by
?
Today, 2:11 PM
|
Responsible plant nutrition requires innovation to improve nutrient use efficiency across diverse agricultural systems. This review highlights nanofertilizers, biofertilizers, and next-generation enhanced-efficiency fertilizers, examining nutrient-release mechanisms, yield impacts, environmental outcomes, and commercialization challenges. However, limited field data and standardized testing hinder progress. To advance fertilizer research, investments in shared protocols, global research networks, and pre-competitive studies are essential to close knowledge gaps, ensure food security, and reduce environmental harm.

|
Scooped by
?
Today, 1:48 PM
|
Over the past two decades, cell-to-cell heterogeneity has garnered increasing attention due to its critical role in both developmental and pathological processes. This growing interest has been driven, in part, by the advancements in live-cell and single-molecule imaging techniques. These techniques have provided mechanistic insights into how processes, transcription in particular, contribute to gene expression noise and, ultimately, cell-to-cell heterogeneity. More recently, however, research has expanded to explore how downstream steps in the central dogma influence gene expression noise. In this review, we mostly examine the impact of transcriptional processes on the generation of gene expression noise but also discuss how post-transcriptional mechanisms modulate noise and its propagation to the protein level. This evaluation emphasizes the need for further investigation into how processes beyond transcription shape gene expression noise, highlighting unanswered questions that remain in the field.

|
Scooped by
?
Today, 1:40 PM
|
Multiple display techniques, including phage display, mRNA display, and ribosome display, have been used to expand the optogenetic toolbox beyond what nature provides. These techniques are most often applied to the development of binding partners that selectively recognize different conformational states of photoswitchable proteins. However, for some targets, in particular the spectrally diverse cyanobacteriochrome (CBCR) GAF domain family, the subtle differences between conformational states pose a significant challenge to discovering highly selective binders. We present an optimized phage display-based protocol designed to effectively capture these subtle changes. This optimized protocol applies high selection pressure by changing the elution method and tightening negative selection, leading to the enrichment of selective binders. Through multiple selection campaigns, we demonstrate the utility of this protocol for identifying highly selective binders.

|
Scooped by
?
Today, 1:04 PM
|
Spermidine finds broad applications across both the nutraceutical and biomedical sectors. In this study, key regulatory genes affecting spermidine synthesis and efficient integration sites were identified to construct a chassis strain for green and sustainable spermidine production. First, the expression of argJ was increased, and the protein SAM2 was mutated to promote the synthesis of spermidine. Second, positional effects were examined in Bacillus amyloliquefaciens. Concurrently, bioinformatics analysis was conducted to uncover transport proteins Blt, YvdR, and Mta, as well as other key genes tcyJ, yxeM, appC, yngA, and orf03307 that affect spermidine synthesis. Ultimately, strain PM13 was constructed through the iterative integration of key genes, achieving a spermidine titer of 396.92 mg/L, 10.34 times higher than strain PM1. Furthermore, xylose fed-batch fermentation increased spermidine titer to 1.69 g/L, setting a new shake flask production record. In conclusion, this study amassed genetic resources and developed an integrated strain for efficient, stable spermidine synthesis.

|
Scooped by
?
Today, 12:37 PM
|
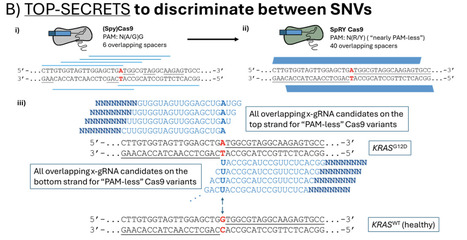
RNA-guided CRISPR nuclease Cas9 cannot reliably differentiate between single nucleotide variations (SNVs) of targeted DNA sequences determined by their guide RNA (gRNA): they typically exhibit similar nuclease activities at any of those variations, unless the variation occurs with specific sequence contexts known as protospacer adjacent motifs (PAMs). Our approach, "TOP-SECRETS," generates gRNA variants that allow Cas9 ribonucleoproteins (RNPs) to reliably discriminate between healthy and disease-associated SNVs outside of PAMs.

|
Scooped by
?
Today, 12:11 PM
|
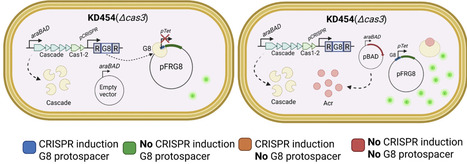
CRISPR-Cas adaptive immunity systems provide defense against mobile genetic elements and are often countered by diverse anti-CRISPR (Acr) proteins. The Type IE CRISPR-Cas of Escherichia coli K12 has been a model for structural and functional studies and is a part of the species' core genome. However, this system is transcriptionally silent, which has fueled questions about its true biological function. To clarify the role of this system in defense, we carried out a census of Acr proteins found in Enterobacterales and identified AcrIE9 as a potent inhibitor of the E. coli K12 Type IE CRISPR-Cas system. While sharing little sequence identity, AcrIE9 proteins from Pseudomonas and Escherichia both interact with the Cas7 subunit of the Cascade complex, thus preventing its binding to DNA. We further show that AcrIE9 is genetically linked to AcrIE10, forming the most widespread anti-CRISPR cluster in Enterobacterales, and this module often co-occurs with a novel HTH-like protein with unusual architecture.
|

|
Scooped by
?
Today, 3:51 PM
|
We demonstrate that the noncanonical amino acid, para-cyanophenylalanine (CNF), when incorporated into metalloproteins, functions as an infrared spectroscopic probe for the redox state of iron sulfur clusters, offering a strategy for determining electron occupancy in the electron transport chains of complex metalloenzymes. We observe a redshift of ≈ 1 - 2 cm-1 in the NC stretching frequency following reduction of spinach ferredoxin modified to contain CNF close to its [2Fe-2S] centre, and this shift is reversed on re-oxidation. We extend this to CNF positioned near to the proximal [4Fe-4S] cluster of the [FeFe] hydrogenase from Desulfovibrio desulfuricans. In combination with a distal [4Fe-4S] cluster and the [4Fe-4S] cluster of the active site ‘H-cluster’ ([4Fe-4S]H), the proximal cluster forms an electron relay connecting the active site to the surface of the protein. Again, we observe a reversible shift in wavenumber for CNF following cluster reduction in either apo-protein (containing the iron-sulfur clusters but lacking the active site) or holo-protein with intact active site, demonstrating general applicability of this approach to studying complex metalloenzymes.

|
Scooped by
?
Today, 3:45 PM
|
Bacterial Mn(II) oxidation is a widespread and ecologically significant process, with Mn oxides playing essential roles in biogeochemical cycling and environmental remediation. However, the environmental cues that initiate this microbial activity remain enigmatic. In this study, a spontaneous mutant was isolated from the previously characterized Arthrobacter strain that exhibited autonomous Mn(II)-oxidizing capacity, in contrast to the parental strain, which required cocultivation with other bacteria to initiate Mn(II) oxidation. Comparative transcriptomic analysis suggested that a gene system encoding an environmental stress-sensing receptor controlled the transcription of boxA, a Mn(II)-oxidizing gene. These findings indicate that two conditions are required for bacterial Mn(II) oxidation: the presence of a Mn(II)-oxidizing gene and an environmentally responsive regulatory system capable of activating its expression. Based on this knowledge, eight additional bacterial strains were isolated that triggered Arthrobacter Mn(II) oxidation. Transcriptomic profiling revealed a robust positive correlation between boxA expression and genes encoding two-component signal transduction systems. Given their widespread distribution (approximately 23% of environmental bacterial genomes harbor Mn(II)-oxidizing gene homologs) and constant exposure to diverse biotic and abiotic stressors, these bacteria are likely to undergo frequent Mn(II) oxidation triggered by microbial interactions or chemical signals. This study provides critical mechanistic insight into the environmental regulation of bacterial Mn(II) oxidation, addressing a longstanding gap in understanding its natural occurrence.

|
Scooped by
?
Today, 3:35 PM
|
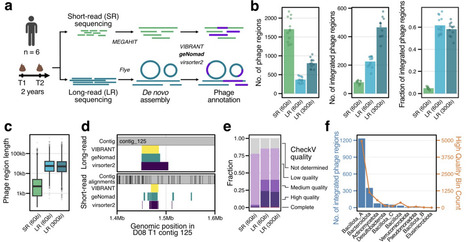
Gut bacteriophages profoundly impact microbial ecology and human health, yet they are greatly understudied. Using deep, long-read bulk metagenomic sequencing, a technique that overcomes fundamental limitations of short-read approaches, we tracked prophage integration dynamics in 12 longitudinal stool samples from six healthy individuals, spanning a two-year timescale. While most prophages remain stably integrated into their host over two years, we discover that ~5% of phages are dynamically gained or lost from persistent bacterial hosts. Within the same sample, we find evidence of population heterogeneity in which identical bacterial hosts with and without a given integrated prophage coexist simultaneously. Furthermore, we demonstrate that phage induction, when detected, occurs predominantly at low levels (1-3x coverage compared to the host region). Interestingly, we identify multiple instances of integration of the same phage into bacteria of different taxonomic families, challenging the dogma that phage are specific to a host of a given species or strain. Lastly, we describe a new class of phages, which we name "IScream phages". These phages co-opt bacterial IS30 transposases to mediate their integration, representing a previously unrecognized form of phage domestication of selfish bacterial elements. Taken together, these findings illuminate fundamental aspects of phage-bacterial dynamics in the human gut microbiome and expand our understanding of the evolutionary mechanisms that drive horizontal gene transfer and microbial genome plasticity in this ecosystem.

|
Scooped by
?
Today, 3:19 PM
|
Protein synthesis in natural cells involves intricate interactions between chemical environments, protein-protein interactions, and protein machinery. Replicating such interactions in artificial and cell-free environments can control the precision of protein synthesis, elucidate complex cellular mechanisms, create synthetic cells, and discover new therapeutics. Yet, creating artificial synthesis environments, particularly for membrane proteins, is challenging due to the poorly defined chemical-protein-lipid interactions. Here, we introduce MEMPLEX (Membrane Protein Learning and Expression), which utilizes machine learning and a fluorescent reporter to rapidly design artificial synthesis environments of membrane proteins. MEMPLEX generates over 20,000 different artificial chemical-protein environments spanning 28 membrane proteins. It captures the interdependent impact of lipid types, chemical environments, chaperone proteins, and protein structures on membrane protein synthesis. As a result, MEMPLEX creates new artificial environments that successfully synthesize membrane proteins of broad interest but previously intractable. In addition, we identify a quantitative metric, based on the hydrophobicity of the membrane-contacting amino acids, that predicts membrane protein synthesis in artificial environments. Our work allows others to rapidly study and resolve the “dark” proteome using predictive generation of artificial chemical-protein environments. Furthermore, the results represent a new frontier in artificial intelligence-guided approaches to creating synthetic environments for protein synthesis. Membrane proteins are notoriously difficult to synthesize. Here, authors introduce MEMPLEX, a high-throughput experimentation platform guided by machine learning that designs artificial cell-free environments to synthesize membrane proteins.

|
Scooped by
?
Today, 2:53 PM
|
The core advantage of acetogen lies in their superior carbon management, achieved by engaging the Wood–Ljungdahl pathway to sequester CO₂ in situ and utilize exogenous CO₂. This advantage can be further exploited by coupling acetogens with other organisms in co-culture, leading to the increasingly explored concept of acetogenic co-cultures. We review and dissect schemes involving the co-fermentation of carbohydrates and exogenous gases, which present unique challenges and opportunities due to the nonlinearity of coculture metabolic networks and complex, often unanticipated, interspecies interactions. The latter suggests that most, if not all, such co-cultures are mutualistic rather than commensalistic, contrary to previous assumptions. We discuss both fundamental and applied concepts, including co-culture stability and methods for quantitatively capturing population dynamics and interspecies interactions.

|
Scooped by
?
Today, 2:38 PM
|
Antimicrobial resistance (AMR) remains a critical challenge in public health and research. AmrProfiler is a comprehensive tool with three specialized modules: identifying acquired AMR genes, resistance-associated mutations, and ribosomal RNA (rRNA) gene mutations across nearly 18 000 bacterial species. By integrating and refining data from established databases, it provides a robust framework for AMR analysis. AmrProfiler is the first to systematically report mutations in rRNA genes, offering in-depth analysis of rRNA copy numbers and mutations—key for identifying potential rRNA-associated resistance mechanisms. Its curated database includes 7600 unique AMR gene entries and over 4300 resistance-related mutations. Supporting genome assemblies in multiple formats, AmrProfiler allows users to customize detection thresholds for AMR genes, mutations, and rRNA analysis. Validation with Acinetobacter baumannii,Escherichia coli,Klebsiella pneumoniae,Staphylococcus aureus, and Staphylococcus epidermidis demonstrated that AmrProfiler accurately identified all AMR genes and mutations reported by other tools while also detecting additional resistance markers and mutations not previously recognized. By bridging AMR genotypes and phenotypes, AmrProfiler provides actionable insights that advance both research and clinical applications in AMR. AmrProfiler is freely available as an open-access web server without login at https://dianalab.e-ce.uth.gr/amrprofiler.

|
Scooped by
?
Today, 2:23 PM
|
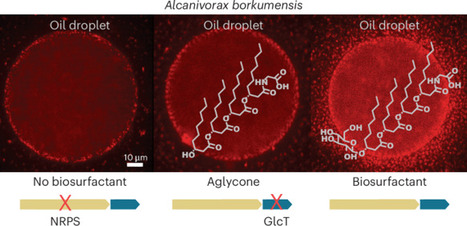
The marine bacterium Alcanivorax borkumensis degrades alkanes derived from phytoplankton, natural hydrocarbon seeps and oil spills. We study the biosynthesis and function of a glycine-glucolipid biosurfactant from A. borkumensis for alkane degradation and identify a gene cluster encoding a nonribosomal peptide synthetase, glycosyltransferase and phosphopantetheinyl transferase. Analyses of A. borkumensis mutants and expression studies reveal that the nonribosomal peptide synthetase catalyzes the synthesis of the aglycone (tetra-d-3-hydroxydecanoyl-glycine) from glycine and d-3-hydroxydecanoyl-CoA, to which a glucose moiety is added by the glycosyltransferase. Deficiency in glycine-glucolipid impairs the ability of mutant cells to attach to the oil–water interface, compromises growth on hexadecane and affects carbon storage. The glycine-glucolipid is essential for biofilm formation on oil droplets and uptake of alkanes. The high incidence of Alcanivorax at oil-polluted sites can in part be explained by the accumulation of the glycine-glucolipid on the cell surface, effectively making the cells themselves act as biosurfactants. Alcanivorax borkumensis is a marine, oil-degrading bacterium. Here, the authors describe the synthesis of a cellular biosurfactant. Loss of the biosurfactant affects attachment of mutant cells to the oil–water interface, highlighting its importance for biofilm formation on oil droplets.

|
Scooped by
?
Today, 1:52 PM
|
Rice agriculture is a major source of atmospheric methane, but current emission inventories are highly uncertain, mostly due to poor rice-specific inundation data. Inversions of atmospheric methane observations can help to better quantify rice emissions but require high-resolution prior information on the location and timing of emissions. Here we use Landsat satellite data at 30 m resolution to map the global monthly distribution of rice paddy fractional areas on a 0.1° × 0.1° (∼10 × 10 km) grid by optimizing an algorithm for flooded vegetation and combining it with a 30 m global cropland database and rice-specific data. We validate this global rice paddy map with an independent US rice database and with seasonal flux measurements from the FLUXNET CH4 network, estimating errors on rice area fraction of 31% on the 0.1° × 0.1° grid and 10% regionally. We combine the rice paddy map with an extensive global data set of emission factors (EFs) per unit of rice paddy area. The resulting Global Rice Paddy Inventory (GRPI) provides methane emission estimates at 0.1° × 0.1° (∼10 × 10 km) spatial resolution and monthly resolution. Our global emission of 39.3 ± 4.7 Tg a−1 for 2022 (best estimate and error standard deviation) is higher than previous inventories that use outdated rice maps and IPCC-recommended EFs now considered to be too low. China is the largest rice emitter in GRPI (8.2 ± 1.0 Tg a−1), followed by India (6.5 ± 1.0 Tg a−1), Bangladesh (5.7 ± 1.2 Tg a−1), Vietnam (5.7 ± 1.0 Tg a−1), and Thailand (4.4 ± 0.9 Tg a−1). These five countries together account for 78% of global total rice emissions. Seasonality of emissions varies considerably between and within individual countries reflecting differences in climate and crop practices. We define a rice methane intensity (methane emission per unit of rice produced) to assess the potential of mitigating methane emission without compromising food security. We find national methane intensities ranging from 10 to 120 kg methane per ton of rice produced (global mean 51) for major rice-growing countries. Countries can achieve low intensities with high-yield cultivars, upland rice agriculture, water management, and organic matter management.

|
Scooped by
?
Today, 1:44 PM
|
Global food shortages and rising antimicrobial resistance require alternatives to antibiotics and agrichemicals for the management of agricultural bacterial pathogens. The phytopathogen Pseudomonas syringae pv. actinidiae (Psa) is the causal agent of kiwifruit canker and is responsible for major agricultural losses. Bacteriophage enzymes present an emerging antimicrobial option. Endolysins possess the ability to cleave peptidoglycan and are effective antimicrobials against gram-positive bacteria. Delivery of endolysins to the peptidoglycan of gram-negatives is impeded by the additional outer membrane. To overcome this barrier, we used VersaTile molecular shuffling to produce Psa-targeting chimeric proteins which were tested for antimicrobial activity. These chimeras consist of endolysins linked by polypeptides to diverse phage proteins mined from Psa phage genomes. A preferential configuration for antibacterial activity was observed for enzymatic domains at the N-terminus and alternative phage proteins at the C-terminus. The lead variant possessed an N-terminal modular endolysin and a C-terminal lipase. Antibacterial activity was enhanced with the addition of the chemical permeabilizers citric acid or EDTA. Mutagenesis of the lipase active site eliminated exogenous antibacterial activity towards Psa. The endolysin-lipase chimera demonstrated specificity towards Psa, illustrating potential as a targeted biocontrol agent. Overall, we generated a chimeric endolysin with exogenous and specific activity towards Psa, the causative agent of kiwifruit canker.

|
Scooped by
?
Today, 1:12 PM
|
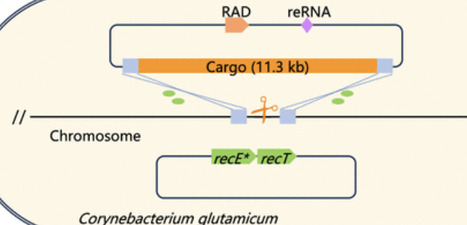
Corynebacterium glutamicum serves as a key microbial chassis for the industrial production of feed and food ingredients. While long DNA fragment insertion technologies have advanced strain engineering capabilities, previous approaches such as utilizing a chromosome-integrated Cas9-RecET system were constrained by a maximum insertion fragment size of 7.5 kb. Through systematic evaluation of Cas9, gRNA, and recombinase expression driven by five distinct promoters and their implementation on 1 or 2 plasmids with compatible replicons (resulting in a total of 17 combinations), we developed an optimized genome editing vector capable of inserting DNA fragments of up to 8.0 kb in C. glutamicum. Parallel implementation of the Cpf1 system also successfully achieved 8.0 kb of DNA insertions. However, the construction of plasmids carrying insertion sequences larger than 8.0 kb was hindered by the plasmid vector capacity. To address this limitation, we screened six smaller RAGATH-associated DNA nucleases, ultimately identifying two with high cleavage activity in C. glutamicum. These nucleases demonstrated superior editing efficiencies compared to both Cas9 and Cpf1, enabling the integration of DNA fragments up to 11.3 kb─surpassing previously reported size limitations for C. glutamicum. These RAGATH-associated DNA nuclease-based systems effectively overcome the previous size constraints for long fragment insertions, thereby advancing metabolic engineering and fundamental research applications.

|
Scooped by
?
Today, 12:47 PM
|
Microbial methylation of arsenic impacts both the toxicity and fate of this environmental contaminant, and is an important component of its biogeochemical cycle. This transformation occurs in flooded paddy fields where soil microorganisms can produce dimethylated arsenic, which causes the straighthead disease in rice. The responsible anaerobic microorganisms have remained elusive because their isolation is laborious, especially as the active methylators cannot be rapidly screened. Here, we introduce a novel approach to specifically target these microorganisms. This approach is based on a high-throughput isolation technique involving microfluidic encapsulation, fluorescence-activated cell sorting (FACS), and biosensor-aided screening of microbial function. Using this method, we isolated two arsenic-methylating anaerobes from a paddy soil. This approach has the potential to rapidly obtain novel isolates. For instance, we show that one isolate actively methylates arsenate (AsV), a previously unknown phenotype in anaerobes.

|
Scooped by
?
Today, 12:28 PM
|
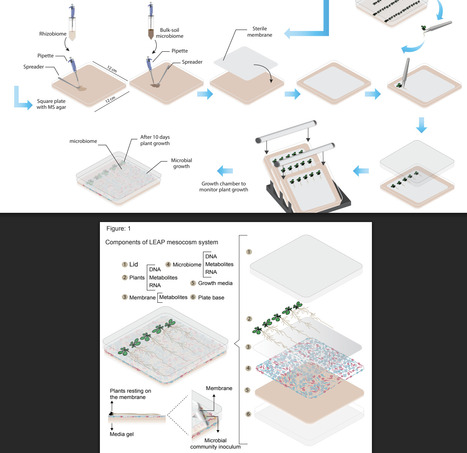
This study introduces an innovative methodology for co-cultivating plants and microbes, employing a membrane-based structure to facilitate their physical separation. The Live-Exudation Assisted Phytobiome Cultromics System (LEAP-CS) has the capability to investigate the complex interaction of plant and soil microbiome under in vitro conditions, offering potential benefits. Subsequent validation and testing can be performed through pot, greenhouse, and field trials. The system can efficiently function as a high-throughput screening tool for assessing plant-microbiome interactions and their associated metabolic signatures. Our phytobiome culturing technique harnesses root exudation from live plants, capitalizing on the membrane's selective separation to prevent direct physical interaction between plant and microbiome components. Consequently, the interaction is solely through chemical-mediated signalling. By employing this method, we can intricately dissect complex plant-microbiome interactions while faithfully maintaining and emulating the contact independent inherent associations prevalent within these phytobiomes. In conclusion, this user-friendly and reproducible 'in-vitro' model holds immense potential for shedding light on the intricate community and metabolic exchange dynamics of plant-microbiome interactions, thus significantly advancing our understanding in this area.
|
 Your new post is loading...
Your new post is loading...




















seelig g, baker d, 2st, better than wang hh system