 Your new post is loading...

|
Scooped by
?
June 6, 4:18 PM
|
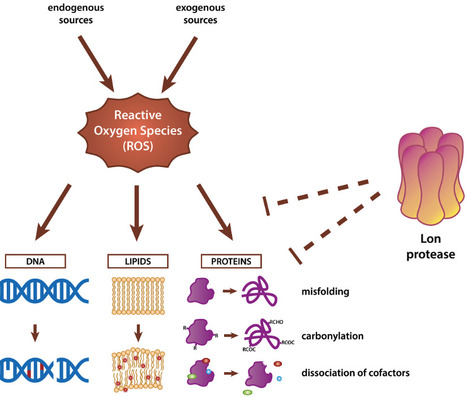
Accumulation of reactive oxygen species (ROS) induces oxidative stress, leading to substantial damage to cellular macromolecules, necessitating efficient protein quality control mechanisms. The Lon protease, a highly conserved ATP-dependent protease, is thought to play a central role in mitigating oxidative stress by targeting damaged and misfolded proteins for degradation. This review examines the role of Lon in oxidative stress responses, including its role in degrading oxidized proteins, regulating antioxidant pathways, and modulating heme and Fe-S cluster homeostasis. We highlight cases of substrate recognition through structural changes and describe situations where Lon activity is further regulated by redox conditions. By synthesizing studies across a range of organisms, we find that despite the clear importance of Lon for oxidative stress tolerance, universal rules for Lon degradation of damaged proteins during this response remain unclear.

|
Scooped by
?
June 6, 3:33 PM
|
Efficient DNA delivery is essential for genetic manipulation of mycobacteria and for dissecting their physiology, pathogenesis, and drug resistance. Although electroporation enables transformation efficiencies exceeding 105 CFU per µg DNA in Mycobacterium smegmatis and Mycobacterium tuberculosis, it remains highly inefficient in many non-tuberculous mycobacteria (NTM), including Mycobacterium abscessus. Here, we discovered that NTM such as M. abscessus exhibit exceptional tolerance to ultra-high electric field strengths, and that hypertonic preconditioning partially protects cells from electroporation-induced damage. Using ultra-high electric field strength (3kV/mm) electroporation, we achieved dramatic improvements in plasmid transformation efficiency—up to 106-fold in M. abscessus, 83-fold in Mycobacterium marinum, and 24-fold in Mycobacterium kansasii—compared to standard conditions (1.25kV/mm). Transformation efficiency was further influenced by the choice of selectable marker. Ultra-high field strength electroporation also markedly enhanced allelic exchange in M. abscessus expressing Che9c RecET recombinases, increasing recovery of gene deletion mutants by over 1,000-fold relative to conventional electroporation. In parallel, oligonucleotide-mediated recombineering for targeted point mutations produced nearly 10,000-fold more mutants under ultra-high field conditions. Together, these findings establish ultra-high field electroporation as a robust, broadly applicable platform for genetic engineering of NTMs. This method substantially enhances transformation efficiency and enables construction of advanced genetic tools—including expression libraries and CRISPRi knockdown libraries—in species that have historically resisted genetic manipulation.

|
Scooped by
?
June 6, 2:10 PM
|
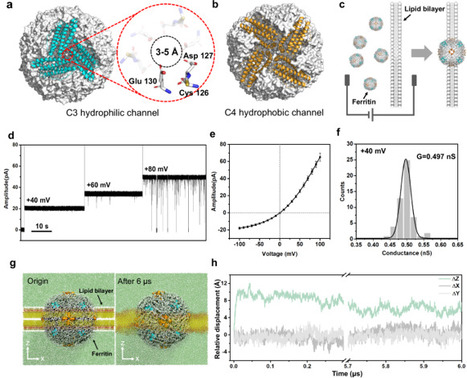
High-resolution nanopore analysis technology relies on the design of novel transmembrane protein platforms. Traditional barrel-shaped protein channels are preferred for constructing nanopore sensors, which may miss protein candidates in non-barrel structures. Here, we demonstrate the globular ferritin displays excellent membrane-insertion capacity and stable transmembrane ionic current owing to its hydrophobic four-fold channels and hydrophilic three-fold channels. The ionic current rectification and voltage-gating characteristics are discovered in single-ferritin ionic current measurement. Notably, the ferritin is used as a nanopore sensor, by which we achieve the high resolution discrimination of L-cysteine, L-homocysteine, and cysteine-containing dipeptides with the assistance of equivalent Cu2+. The mechanistic studies by multiple controlled experiments and quantum mechanics/all-atom/coarse-grained multiscale MD simulations reveal that analytes are synergistically captured by His114, Cys126, and Glu130 within C3 channel, causing the current blockage signals. The promising ferritin nanopore sensor provides a guide to discovering new protein nanopores without shape restrictions. Traditional nanopore sensors use barrel-shaped protein channels. Here, the authors report on a study into the use of globular protein, ferritin, as a nanopore sensor, demonstrating membrane insertion and sensor application, showing the potential of non-barrel-shaped proteins for nanopore sensing.

|
Scooped by
?
June 6, 1:30 PM
|
Engineering synthetic consortia to perform distributed functions requires robust quorum sensing (QS) systems to facilitate communication between cells. However, the current QS toolbox lacks standardized implementations, which are particularly valuable for use in bacteria beyond the model species Escherichia coli. We developed a set of three QS systems encompassing both sender and receiver modules, constructed using backbones from the SEVA (Standard European Vector Architecture) plasmid collection. This increases versatility, allowing plasmid features like the origin of replication or antibiotic marker to be easily swapped. The systems were characterized using the synthetic biology chassis Pseudomonas putida. We first tested individual modules, then combined sender and receiver modules in the same host, and finally assessed the performance across separate cells to evaluate consortia dynamics. Alongside the QS set, we provide mathematical models and rate parameters to support the design efforts. Together, these tools advance the engineering of robust and predictable multicellular functions.

|
Scooped by
?
June 6, 2:03 AM
|
Herbicide resistance (HR) is fundamental for sustainable agriculture as global food security increasingly relies on efficient and eco-friendly weed management. Recent advances in CRISPR/dCas9-based epigenome editing offer a promising, non-genetic approach by precisely targeting regulatory regions of genes involved in herbicide sensitivity and detoxification. While CRISPR/Cas9 has successfully been used to develop HR crops, CRISPR/dCas9 remains underexplored in this field. We propose that CRISPR/dCas9-driven epigenome editing could enable time- and tissue-specific control of gene expression, allowing for the introduction of heritable HR traits without altering DNA sequences. This innovative approach could transform sustainable HR development, offering a powerful solution to enhance agricultural resilience and food security while aligning with eco-friendly weed management strategies.

|
Scooped by
?
June 6, 1:38 AM
|
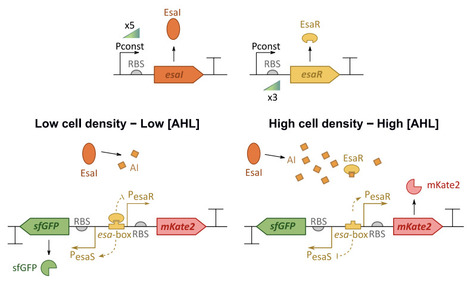
Quorum sensing-based genetic circuits are gaining traction in synthetic biology as they link population-level behavior to individual cell responses. However, tuning these circuits remains challenging due to complex dynamics, particularly during the 'Learn' phase of the Design-Build-Test-Learn (DBTL) cycle. To accelerate this process, we developed a mathematical model to predict how varying expression levels of the transcription factor and synthase affect the response of the EsaI/EsaR quorum sensing system. A strain library was constructed, and experimental data were used to optimize the model. The final model could successfully differentiate between the effects of these expression levels on the response of the bidirectional promoter. It allowed visualization of all potential system outcomes and emphasized the transcription factor's critical role in tuning the circuit. This model offers a valuable tool for fine-tuning EsaI/EsaR-based systems for synthetic biology applications. Moreover, given the homology within the LuxR-family quorum sensing systems, this modelling approach may serve as a foundation for model-based tuning of other quorum sensing systems.

|
Scooped by
?
June 6, 1:31 AM
|
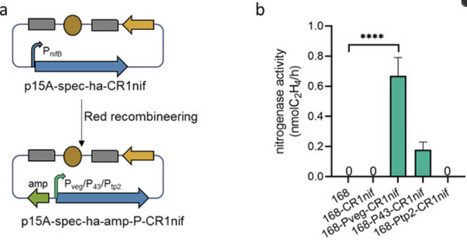
Microbially mediated biological nitrogen fixation is pivotal to sustainable agricultural development. However, optimizing nitrogenase activity in native biological nitrogen-fixing bacteria has been hindered by the complexities of genetic manipulation. Heterologous expression has served as a foundational strategy for engineering next-generation nitrogen-fixing microbial agents. In this study, genomic analysis of Paenibacillus polymyxa CR1 revealed an 11 kb nitrogen-fixing (nif) gene cluster. The nif cluster was first synthesized and then assembled using ExoCET technology and finally integrated into the genome of Bacillus subtilis 168 via double-exchange recombination. RT-PCR confirmed the transcription of the nif cluster; however, no nitrogenase activity was detected in the acetylene reduction assay. A promoter replacement strategy (replacing the native promoter with Pveg) enabled B. subtilis to produce active nitrogenase. However, stronger promoters—namely, P43 and Ptp2—did not further enhance nitrogenase activity. This demonstrates that promoter selection requires balancing transcriptional strength with systemic compatibility, particularly for metalloenzymes demanding precise cofactor assembly. This is the first report describing the heterologous expression of the nif gene cluster in B. subtilis, establishing a foundation for engineering high-efficiency nitrogen-fixing biofertilizers.

|
Scooped by
?
June 6, 1:25 AM
|
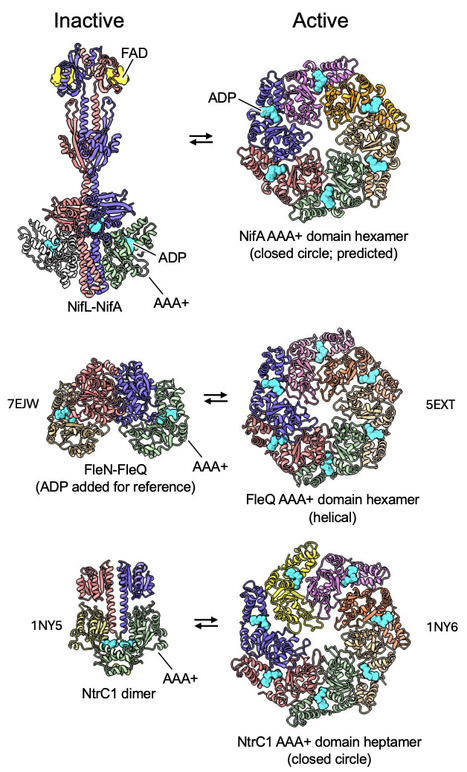
Understanding the molecular basis of regulated nitrogen (N2) fixation is essential for engineering N2-fixing bacteria that fulfill the demand of crop plants for fixed nitrogen, reducing our reliance on synthetic nitrogen fertilizers. In Azotobacter vinelandii and many other members of Proteobacteria, the two-component system NifL-NifA controls the expression of nif genes that encode the nitrogen fixation machinery. The NifL-NifA system evolved the ability to integrate several environmental cues, such as oxygen, nitrogen, and carbon availability. The nitrogen fixation machinery is thereby only activated under strictly favorable conditions, enabling diazotrophs to thrive in competitive environments. Whilst genetic and biochemical studies have enlightened our understanding of how NifL represses NifA, the molecular basis of NifA sequestration by NifL depends on structural information on their interaction. Here, we present mechanistic insights into how nitrogen fixation is regulated by combining biochemical and genetic approaches with a low-resolution cryo-EM map of the oxidized NifL-NifA complex. Our findings define the interaction surface between NifL and NifA and reveal how this interaction can be manipulated to generate bacterial strains with increased nitrogen fixation rates able to secrete surplus nitrogen outside the cell, a crucial step in engineering improved nitrogen delivery to crop plants.

|
Scooped by
?
June 6, 1:18 AM
|
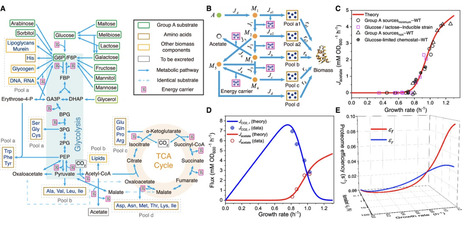
A classic problem in metabolism is that fast-proliferating cells use seemingly wasteful fermentation for energy biogenesis in the presence of sufficient oxygen. This counterintuitive phenomenon, known as overflow metabolism or the Warburg effect, is universal across various organisms. Despite extensive research, its origin and function remain unclear. Here, we show that overflow metabolism can be understood through growth optimization combined with cell heterogeneity. A model of optimal protein allocation, coupled with heterogeneity in enzyme catalytic rates among cells, quantitatively explains why and how cells choose between respiration and fermentation under different nutrient conditions. Our model quantitatively illustrates the growth rate dependence of fermentation flux and enzyme allocation under various perturbations and is fully validated by experimental results in Escherichia coli. Our work provides a quantitative explanation for the Crabtree effect in yeast and the Warburg effect in cancer cells and can be broadly used to address heterogeneity-related challenges in metabolism.

|
Scooped by
?
June 6, 12:48 AM
|
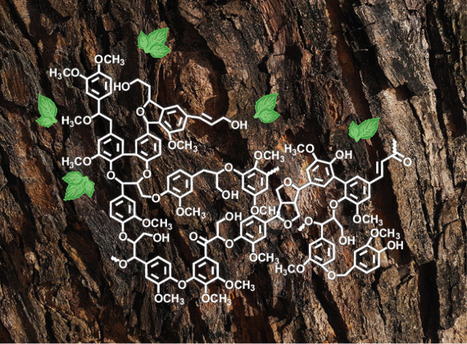
Moses Dike and Shudipto Konika Dishari explore lignin’s historic journey alongside human civilization and showcase its game-changing potential to drive sustainability without compromising performance.

|
Scooped by
?
June 6, 12:21 AM
|
Converting lignocellulose into bioelectricity through a bioelectrocatalytic system (BES) has emerged as a promising approach to addressing environmental pollution and energy regeneration challenges. However, practical application of BES is significantly constrained by the fact that the electroactive biocatalyst Shewanella oneidensis lacks the essential metabolic pathways and enzymes required for utilizing lignocellulose for cell growth and power generation. Here, to realize clean electricity production from lignocellulose hydrolysate, an artificial microbial consortium comprising S. oneidensis, Lactococcus lactis, and Bacillus subtilis was developed. In this consortium, L. lactis is responsible for converting glucose into lactate; B. subtilis metabolizes glucose and xylose into riboflavin; and S. oneidensis then employs lactate as an electron donor and riboflavin as an electron shuttle to facilitate electricity generation. Subsequently, to increase substrate conversion efficiency of the microbial consortium, three key genes codY, ribA, and dld encoding lactate dehydrogenase, GTP cyclohydrolase, and d-lactate dehydrogenase, were expressed in L. lactis, B. subtilis, and S. oneidensis, respectively, which accelerated glucose-to-lactate conversion, riboflavin synthesis, and lactate metabolism. Also, to accelerate the extracellular electron transfer (EET) capacity of the microbial consortium, the cyc2 gene from Acidithiobacillus ferrooxidans encoding the outer membrane c-type cytochrome was further expressed in S. oneidensis. Finally, to further enhance the interfacial EET capability of the microbial consortium, a 3D microbiota biohybrid system S7L1B1@CF&GO consisting of carbon felts and graphene oxide was developed to reduce the internal resistance of BES. The results showed that the artificial biohybrid system could obtain a maximum power density of ∼739.40 mW m–2 using lignocellulosic hydrolysate as the carbon source. This system expands the range of carbon sources available to S. oneidensis for efficient power generation from the lignocellulosic hydrolysate.

|
Scooped by
?
June 5, 11:49 PM
|
Surface sensing is a key aspect of the early stage of biofilm formation. For Pseudomonas aeruginosa PA14, the type IV pili (T4P), the T4P alignment complex, and PilY1 were shown to play a key role in c-di-GMP signaling upon surface contact. The role of the flagellar machinery in surface sensing is less well understood for P. aeruginosa. Here, we show, consistent with findings from other groups, that a mutation in the gene encoding the flagellar hook protein (ΔflgK) or flagellin (ΔfliC) results in a strain that overproduces the Pel exopolysaccharide (EPS) with a concomitant increase in c-di-GMP levels. We use a candidate gene approach and genetic screens, combined with phenotypic assays, to identify key roles for the MotAB and MotCD stators and the FliG protein, a component of the flagellar switch complex, in stimulating the surface-dependent, increased c-di-GMP level noted for these flagellar mutants. These findings are consistent with previous studies showing a role for the stators in surface sensing. We also show that mutations in the genes coding for the DGCs SadC and RoeA, as well as SadB, a protein involved in early surface colonization, abrogate the increased c-d-GMP-related phenotypes of the ΔflgK mutant. Together, these data indicate that bacteria monitor the status of flagellar synthesis and function during surface sensing as a mechanism to trigger the biofilm program.

|
Scooped by
?
June 5, 11:33 PM
|
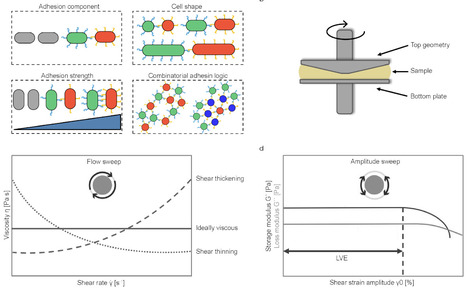
Engineered living materials (ELM) at the multicelluar level represent an innovation that promises programmable properties for biomedical, environmental, and consumer applications. However, the rational tuning of the mechanical properties of such ELMs from first principles remains a challenge. Here we use synthetic cell-cell adhesins to systematically characterize how rheological and viscoelastic properties of multicellular materials made from living bacteria can be tuned via adhesin strength, cell size and shape, and adhesion logic. We confirmed that the previous results obtained for non-living materials also apply to bacterial ELMs. Additionally, the incorporation of synthetic adhesins, combined with the adaptability of bacterial cells in modifying various cellular parameters, now enables novel and precise control over material properties. Furthermore, we demonstrate that rheology is a powerful tool for actively shaping the microscopic structure of ELMs, enabling control over cell aggregation and particle rearrangement, a key feature for complex material design. These results deepen our understanding of tuning the viscoelastic properties and fine structure of ELMs for applications like bioprinting and microbial consortia design including natural systems.
|

|
Scooped by
?
June 6, 3:59 PM
|
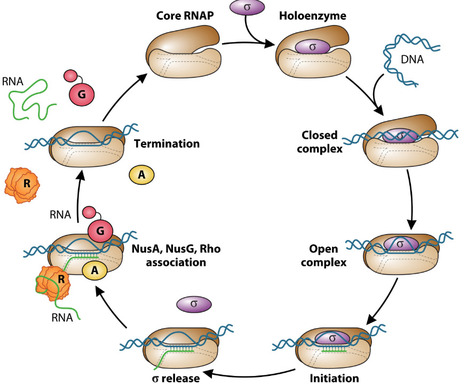
Transcription by RNA polymerase is punctuated by transient pausing events. Pausing provides additional time for proper RNA folding and binding of regulatory factors to the paused transcription elongation complex or the nascent RNA. Depending on the organism and the genomic context, the general transcription elongation factors NusA and NusG stimulate or suppress pausing. Both Escherichia coli and Bacillus subtilis NusA stimulate pausing in vitro, while the genome-wide role of NusA on pausing has only been examined in B. subtilis. NusG-dependent pausing was identified throughout the B. subtilis genome, and in several instances, these pauses were shown to regulate the expression of the downstream gene(s). This pro-pausing activity was also observed for Mycobacterium tuberculosis NusG. In contrast, E. coli NusG functions as an anti-pausing factor by suppressing pausing throughout the genome. These differences in the function of NusG highlight the importance of studying fundamental processes in a variety of bacterial species. This review will highlight recent advances gained by the ability to identify pauses genome-wide that are either stimulated or suppressed by these two conserved transcription elongation factors.

|
Scooped by
?
June 6, 3:09 PM
|
Agricultural soils are an important source of nitrous oxide (N2O), which has greenhouse and ozone-depleting effects. Bradyrhizobium ottawaense SG09 is a nitrogen-fixing rhizobium with high N2O-reducing activity. Rhizobia form symbiotic nodules in leguminous plants. The initial physical attachment of bacteria to plant roots is a critical step in the establishment of symbiotic interactions. In this study, we performed microscopic analysis using DsRed-expressing B. ottawaense SG09. We revealed that B. ottawaense SG09 attached to both the root surface and root hairs via single cellular poles. This polar attachment was observed not only to the symbiotic host soybean, but also to non-leguminous plants, such as Arabidopsis, rice, corn, and wheat. We identified and analyzed the unipolar polysaccharide (upp) gene cluster, which is proposed to be involved in polar attachment of rhizobia, in the genome of B. ottawaense SG09. We established an Arabidopsis-based interaction assay and demonstrated that uppC and uppE play a critical role in attachment to both the root surface and root hairs.

|
Scooped by
?
June 6, 1:44 PM
|
Reducing the prevalence of phytopathogens and their impact on crops is essential to reach sustainable agriculture goals. Synthetic pesticides have been commonly used to control crop disease but are now strongly linked to disease resistance, environmental pollution, depletion of soil biodiversity, and bioaccumulation, leading to adverse effects on human health. As a alternative, the prolific Trichoderma genus has been studied for its biocontrol properties, as well as its ability to promote plant growth and increase nutrient uptake. This is done through various mechanisms, one of which is the production of bioactive natural products with high chemical diversity. These include terpenoids, alkaloids, non-ribosomal peptides, polyketides and RiPPs. One of the most studied examples is 6-pentyl-2H-pyran-2-one, a volatile organic polyketide, which induces systemic acquired resistance, morphogenesis, and natural product biosynthesis in plants. Methods for culturing Trichoderma spp., isolating and characterising unique bioactive metabolites are discussed here, with an emphasis on dereplication strategies using metabolomics to optimise discovery. In addition, the role of genome mining for the study of natural product biosynthesis in Trichoderma, and more generally, filamentous fungi is discussed. Examples of bioinformatics tools available to date are listed here with applications in Trichoderma and other ascomycetes. New advances in genome engineering in Trichoderma are also detailed, providing insights into available strategies for the validation of biosynthetic gene clusters identified using genome mining. Finally, the use of a combination of omics approaches, namely metabologenomics, is presented as a growing field for natural product discovery in fungi.

|
Scooped by
?
June 6, 1:23 PM
|
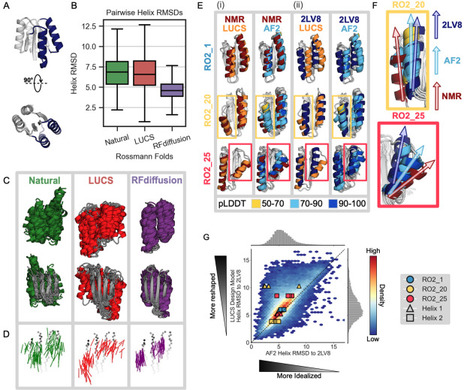
Nature uses structural variations on protein folds to fine-tune the geometries of proteins for diverse functions, yet deep learning-based de novo protein design methods generate highly regular, idealized protein fold geometries that fail to capture natural diversity. Here, using physics-based design methods, we generated and experimentally validated a dataset of 5,996 stable, de novo designed proteins with diverse non-ideal geometries. We show that deep learning-based structure prediction methods applied to this set have a systematic bias towards idealized geometries. To address this problem, we present a fine-tuned version of Alphafold2 that is capable of recapitulating geometric diversity and generalizes to a new dataset of thousands of geometrically diverse de novo proteins from 5 fold families unseen in fine-tuning. Our results suggest that current deep learning-based structure prediction methods do not capture some of the physics that underlie the specific conformational preferences of proteins designed de novo and observed in nature. Ultimately, approaches such as ours and further informative datasets should lead to improved models that reflect more of the physical principles of atomic packing and hydrogen bonding interactions and enable improved generalization to more challenging design problems.

|
Scooped by
?
June 6, 1:55 AM
|
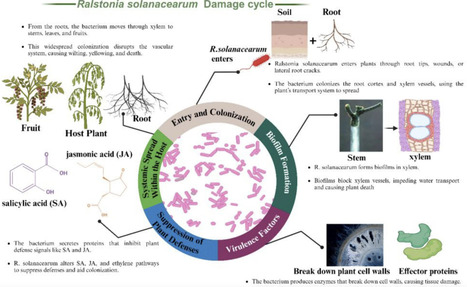
Ralstonia solanacearum, a Gram-negative bacterium, is the causative agent of bacterial wilt, a devastating disease affecting a wide range of economically important crops worldwide. This study explores the dynamic interactions between Ralstonia solanacearum and its host plants, emphasizing key mechanisms underlying infection and host response. The pathogen initiates infection through root wounds or natural openings, rapidly colonizing xylem vessels where it forms biofilms that disrupt water and nutrient transport. Its virulence is driven by cell wall-degrading enzymes and effector proteins delivered via a Type III secretion system, which subvert plant immune responses and facilitate systemic spread. In turn, host plants activate hormonal and stress-related defense pathways, though these are often manipulated by the pathogen, leading to disease progression and reduced productivity. This review highlights critical gaps in our understanding of molecular host-pathogen interactions and the role of environmental conditions in disease development. Addressing these gaps is vital for improving management strategies, with breeding for resistance and advanced biotechnological tools offering promising solutions to combat bacterial wilt and support sustainable agriculture. Future research should focus on leveraging genetic insights to enhance host resistance, employing advanced biotechnological tools to develop crop varieties with enhanced resistance to Ralstonia solanacearum, thereby promoting sustainable agriculture and strengthening global food security.

|
Scooped by
?
June 6, 1:34 AM
|
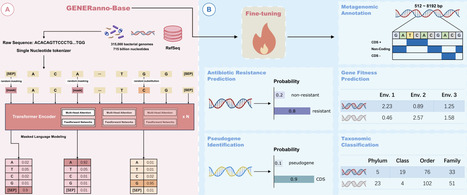
The rapid growth of genomic and metagenomic data has underscored the pressing need for advanced computational tools capable of deciphering complex biological sequences. In this study, we introduce GENERanno, a compact yet powerful genomic foundation model specifically optimized for metagenomic annotation. Trained on an extensive dataset comprising 715 billion base pairs (bp) of prokaryotic DNA, GENERanno employs a transformer encoder architecture with 500 million parameters, enabling bidirectional attention over sequences up to 8192 nucleotides at single-nucleotide resolution. This design addresses key limitations of existing methods, including the inability of traditional Hidden Markov Models (HMMs) to handle fragmented DNA sequences, as well as the suboptimal tokenization schemes of current genomic foundation models that compromise fine-grained analysis. To evaluate the model performance, we curated the Prokaryotic Gener Tasks, a biologically meaningful benchmark encompassing gene fitness prediction, antibiotic resistance prediction, gene classification, and taxonomic classification. Across these tasks, GENERanno consistently outperforms its counterparts, establishing itself as a leading genomic foundation model in the prokaryotic domain. For metagenomic annotation, GENERanno achieves superior accuracy compared to traditional HMM-based methods (e.g., GLIMMER3, GeneMarkS2, Prodigal) and recent LLM-based approaches (e.g., GeneLM), while demonstrating exceptional generalization ability on archaeal genomes. Notably, GENERanno pioneers the prediction of pseudogenes based solely on sequence data, leveraging its contextual understanding to differentiate non-functional sequences from active coding regions. Overall, GENERanno represents a significant advancement in genomic foundation modeling, bridging the gap between large-scale sequence analysis and fine-grained biological insights. By providing a versatile tool for metagenomic annotation and broader genomic exploration, this work lays the groundwork for future research in functional genomics and related fields. https://github.com/GenerTeam/GENERanno

|
Scooped by
?
June 6, 1:29 AM
|
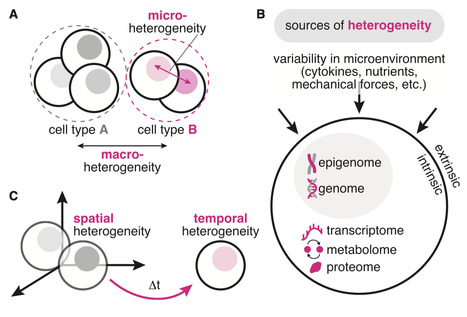
Systems biology offers a view of the cell as an input-output device: a biochemical network (or cellular “processor”) that interprets cues from the microenvironment to drive cell fate. Advancements in single-cell technologies are unlocking the cellular black box, revealing heterogeneity in seemingly homogeneous cell populations. But are these differences technical variability or biology? In this review, we explore this question through a systems biology lens, offering a framework for conceptualizing heterogeneity from the cell’s perspective and summarizing systems and synthetic biology tools for capturing heterogeneity. While cellular inputs shape the probability of attaining particular fates, each cell spins a stochastic “wheel of fate.” Applying this framework, we explore heterogeneity in two case studies: human pluripotent stem cell (hPSC) culture and beta cell differentiation. Looking forward, we discuss how a systems approach to heterogeneity may enable more predictable outcomes in stem cell research, with broad implications for developmental biology and regenerative medicine.

|
Scooped by
?
June 6, 1:21 AM
|
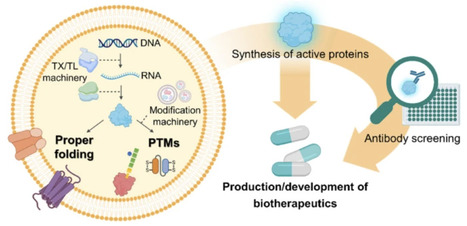
The convergence of cell-free protein synthesis (CFPS) and vesicle-based delivery platforms presents a promising avenue for therapeutic development. The open environment of CFPS offers precise control over protein synthesis by enabling the modulation of synthetic conditions. Additionally, vesicle-based platforms provide enhanced stability, bioavailability, and targeted delivery. This synergy facilitates the efficient production of complex proteins—including membrane proteins, antibody fragments, and proteins requiring post-translational modifications (PTMs)—and supports novel drug delivery strategies. While existing reviews have covered synthetic cells and biomanufacturing broadly, a dedicated analysis of CFPS system-containing vesicles (CFVs) for therapeutic applications remains absent from the literature. This review addresses this knowledge gap by providing a comprehensive examination of CFVs, highlighting their potential as programmable drug delivery platforms through the integration of genetic circuits. It emphasizes the advantages of CFPS over traditional cell-based approaches and explores the synergistic benefits of combining CFPS with various vesicle systems. These systems offer dynamic control over therapeutic protein production and targeted delivery, enabling precise responses to specific signals in complex environments. Although challenges such as low protein yield and imperfect targeting remain, potential optimization strategies are discussed. This analysis highlights the significant potential of integrating CFPS and vesicle-based delivery to advance biomanufacturing, therapeutic development, and synthetic cell systems, thereby opening new avenues in medicine and healthcare.

|
Scooped by
?
June 6, 1:10 AM
|
RNA-guided CRISPR-Cas nucleases are widely used as versatile genome-engineering tools. Among the diverse CRISPR-Cas effectors, CRISPR-Casλ—also referred to as Cas12n—is a recently identified miniature type V nuclease encoded in phage genomes. Given its demonstrated nuclease activity in both mammalian and plant cells, Casλ has emerged as a promising candidate for genome-editing applications. However, the precise molecular mechanisms of Casλ family enzymes remain poorly understood. In this study, we report the identification and detailed biochemical and structural characterizations of CRISPR-Casλ2. The cryo-electron microscopy structures of Casλ2 in five different functional states unveiled the dynamic domain rearrangements during its activation. Our biochemical analyses indicated that Casλ2 processes its precursor crRNA to a mature crRNA using the RuvC active site through a unique ruler mechanism, in which Casλ2 defines the spacer length of the mature crRNA. Furthermore, structural comparisons of Casλ2 with Casλ1 and CasΦ highlighted the diversity and conservation of phage-encoded type V CRISPR-Cas enzymes. Collectively, our findings augment the mechanistic understanding of diverse CRISPR-Cas nucleases and establish a framework for rational engineering of the CRISPR-Casλ-based genome-editing platform. In this study, the authors report the identification, biochemical characterization and cryo-electron microscopy structure of a phage-encoded miniature type V CRISPR-Cas nuclease in different functional states. This work provides mechanistic insight into CRISPR-Cas12 nucleases and a solid framework for future rational engineering.

|
Scooped by
?
June 6, 12:47 AM
|
Protein secretion in yeast is a complex, multistep process heavily reliant on signal sequences to guide recombinant proteins through the secretory pathway. Among these, the mating factor alpha (MFα) signal sequence from Saccharomyces cerevisiae has emerged as a powerful tool for enhancing the extracellular production of heterologous proteins. This review provides a comprehensive overview of the MFα signal sequence, tracing its historical development and role in advancing our understanding of protein secretion mechanisms, including co- and post-translational secretory pathways. We highlight key studies focused on optimizing the MFα signal sequence for improved secretion efficiency, leading to the development of several highly effective variants. These optimized sequences have significantly increased recombinant protein yield and quality, with notable implications for both research and industrial applications. Additionally, we explore the challenges of MFα-based secretion, including issues of missorting, incorrect processing, and aggregation in the endoplasmic reticulum (ER). We discuss emerging strategies to overcome these bottlenecks, such as fusion with alternative signal sequences and strain engineering. Finally, the review highlights current efforts to develop more robust signal peptides, and underscores the importance of continued innovation in protein secretion systems to meet the growing demand for high-quality recombinant proteins in biotechnological and therapeutic applications.

|
Scooped by
?
June 6, 12:12 AM
|
Glycans cover the surfaces of all cells, where they are poised to mediate a two-way discourse between bacteria and host cells. In some instances, glycan-mediated interactions foster a symbiotic state, and in others, they tip the balance toward disease. Chemical biology approaches have begun to reveal the roles of glycans in host-bacteria interactions and provide novel pathways to modulate these interactions. Here, we highlight recent advances in the development and application of chemical biology tools to delineate the roles of glycans in bacterial adhesion, bacterial evasion of the host immune system, host recognition of bacterial cells, and endogenous mechanisms to maintain symbiosis. Further, we present glycan-based strategies to disrupt host-pathogen interactions and to promote the growth of beneficial bacteria.

|
Scooped by
?
June 5, 11:33 PM
|
Peer review is the process by which the quality of scholarly work is assessed prior to being published, presented, or funded. The consequences of flawed research entering the public domain in the “post-truth” era highlight the need to improve peer review quality, which we believe can be achieved by standardizing training. Here, we aim to enhance the quality of published literature by presenting a systematic guide to train new reviewers (and aid experienced ones) in the art of peer reviewing the rigor of scientific manuscripts.
|
 Your new post is loading...
Your new post is loading...
















mode of regulation, riboswitch, thermosensor, temperature