 Your new post is loading...

|
Scooped by
?
Today, 1:55 AM
|
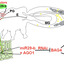
miR29-b,along with potentially other effectors and elicitors, is synthesized in the primary salivary glands (PGs) of Bemisiatabaciand secreted into phloem cells (Ps) via an exosome-mediated process (green circle). During feeding, miR29-b is released with saliva through the salivary duct (red line) originating from the salivary glands. Once inside the plant, miR29-b interacts with AGO1, triggering gene silencing of the plant BAG4 gene, which typically activates defense responses linked to the jasmonic acid (JA) and salicylic acid (SA) pathways. This suppression weakens the plant’s defense mechanisms associated with these phytohormones. The green line represents the food canal, while the yellow circles indicate bacterial endosymbionts involved in insect–plant interactions. AG, accessory salivary glands; E, esophagus, MG, midgut; HG, hindgut; EG, eggs.

|
Scooped by
?
Today, 1:45 AM
|
Cytochrome P450 enzymes (P450s) are versatile biocatalysts with applications spanning pharmaceutical development and natural product biosynthesis. A critical bottleneck in P450-mediated reactions is the electron transfer process, which often limits catalytic efficiency and promotes uncoupling events leading to reactive oxygen species (ROS) formation. This review comprehensively examines recent protein engineering strategies aimed at enhancing electron transfer efficiency in P450 systems. We explore the design and application of different fusion constructs, which improve proximity between the P450 enzyme and its redox partners (RPs), as well as scaffold-mediated protein assembly, enabling precise spatial organization of P450s and RPs. Furthermore, we discuss targeted modifications at the P450-RP interaction interface and optimization of electron transfer pathways through site-directed mutagenesis and directed evolution. These strategies enhance catalytic activity, improve coupling efficiency, and reduce ROS formation. Finally, we address the remaining challenges in understanding and engineering P450 electron transfer, and discuss the future directions, emphasizing the need for advanced computational modeling, structural characterization, and integration of synthetic and systems biology approaches.

|
Scooped by
?
Today, 1:17 AM
|
Recent studies underscore the significant role of RNA secondary structures in various biological and pathological processes. Structural conservation can reveal homologies undetectable by sequence analysis alone, making accurate prediction and comparison of RNA secondary structures crucial. The BEAGLE algorithm enables pairwise alignments of RNA secondary structures through dynamic programming, leveraging the BEAR encoding for RNA secondary structures representation. Initially, BEAGLE was designed to perform pairwise alignments of user-provided RNAs or against a limited number of datasets. We now introduce BEAGLE 2.0, a web server designed to facilitate the search for structural similarities between user-provided RNA molecules and an expanded collection of RNA secondary structure datasets. These datasets include structures derived from SHAPE experiments in Homo sapiens, Mus musculus, Danio rerio, Escherichia coli, Bacillus subtilis, and various viruses, including SARS-CoV-2. It also incorporates predicted structures from the NONCODE database for a wide range of animals and plants, as well as a dataset of structures based on constraints derived from conserved positions within the families present in Rfam. Users can input RNA sequences or a combination of sequences and secondary structures in either dot-bracket or BEAR format. BEAGLE 2.0 outputs pairwise alignments with measures of structural similarity and statistical significance. Additionally, it offers a visual representation of the secondary structures, with structural elements highlighted in different colors. Overall, BEAGLE 2.0 enables searches in RNA structure datasets leveraging experimentally supported data, to identify structural similarities in RNAs of interest. BEAGLE 2.0 is available at https://beagle2.bio.uniroma2.it.

|
Scooped by
?
Today, 12:59 AM
|
Synthetic genetic materials, particularly those in genetically modified organisms (GMO) deployed into complex environments, necessitate robust postmarket surveillance for continuous monitoring of both the materials and their applications throughout their lifecycle. Here, we introduce novel-coded genomic material for a blind mutation test that evaluates mutagenesis in synthetic genomic sequences without requiring direct sequence comparison. This test utilizes a Genome-Digest, which is embedded within essential genes, establishing mathematical correlation between the nucleotide sequence and codon order. This novel design allows for independent assessment of mutations by decoding the nucleotide sequence, thereby eliminating the need for reference sequences or extensive bioinformatic analysis. Furthermore, the test has the capability to analyze mixed genomic materials from a single sample and can be extended to the pooled testing of multiple samples as well. Building on this framework, we propose the ‘Genome-ShockWatch’ methodology. In proof-of-concept trials, it successfully detected mutations that exceeded a predefined threshold in long-read sequencing data from a yogurt sample containing Genome-Digest encoded Nissle 1917 E. coli cells and naturally occurring probiotic bacteria. Consequently, the Genome-Digest system provides a robust foundation for the routine surveillance and management of GMOs and related synthetic products, ensuring their safety and efficacy in diverse environmental contexts.

|
Scooped by
?
Today, 12:18 AM
|
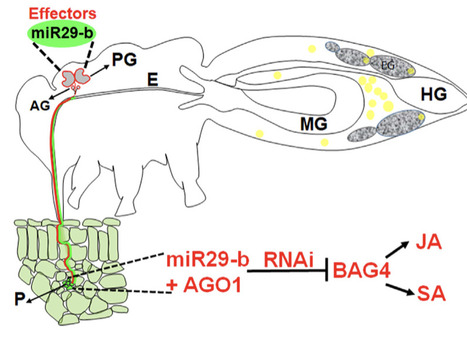
Continuous cropping (CC) obstacles are among the primary factors limiting the development of global agriculture. Although most plants are negatively affected by barriers that develop with CC, they may also overcome obstacles by altering the soil biological and chemical environment to favor plant growth. In this study, we investigated the mechanism by which plants recruit beneficial microorganisms using root exudates to alleviate obstacles in a 10- year CC potato system. On day 20 after potato emergence in a CC system, soil microorganisms promoted an increase in adventitious root (AR) numbers by increasing indole-3-acetic acid (IAA) content in rhizosphere soil. Analysis of rhizosphere bacterial communities using 16S rRNA sequencing revealed that CC altered community structure, with increased abundance of Pantoea sp. MCC16. Irrigation with root exudates from CC potato significantly increased AR numbers and Pantoea sp. MCC16 abundance. According to untargeted metabolomic analysis, nobiletin was identified as promoting Pantoea sp. MCC16 colonization in the rhizosphere. Last, application of nobiletin or Pantoea sp. MCC16 significantly increased the yield of CC potatoes. Thus, CC plants can actively secrete unique metabolites nobiletin to recruit Pantoea sp. MCC16, a high IAA producer, to help plants recover functional traits and mitigate CC obstacles.

|
Scooped by
?
Today, 12:00 AM
|
OT-Mation is an open-source Python script designed to automate the programming of OT-2 liquid-handling robots, making combinatorial experiments more accessible to researchers. By parsing user-defined CSV files containing information on labware, reagents, pipettes, and experimental design, OT-Mation generates a bespoke Python script compatible with the OT-2 system. OT-Mation enhances reproducibility, reduces human error, and streamlines workflows, making it a valuable addition to any laboratory utilizing OT-2 robotics for liquid handling. While OT-Mation can be used for setting up any type of experiment on the OT-2, its real utility lies in making the connection between multifactorial experimental design software outputs (i.e. design of experiments arrays) and liquid-handling robot executable code. As such, OT-Mation helps bridge the gap between code-based flexibility and user-friendly operation, allowing researchers with limited programming skills to design and execute complex experiments efficiently. opentron

|
Scooped by
?
May 12, 11:49 PM
|
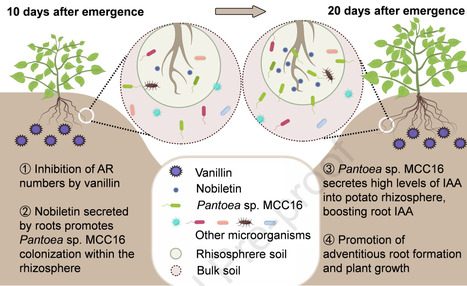
Bacteria rely on an arsenal of weapons to challenge their opponents in highly competitive environments. To specifically counter closely related bacteria, specialized weapons with a narrow activity spectrum are deployed, particularly contractile phage tail-like particles or R-tailocins. Their production leads to the lysis of the producing cells, indicating that their expression must be carefully orchestrated so that only a small percentage of cells produce R-tailocins for the benefit of the entire population. In this study, we set out to better understand how the production of these phage tail-like weapons is regulated in environmental pseudomonads using the competitive plant root colonizer and environmental model strain Pseudomonas protegens CHA0. Using an RNA sequencing (RNA-seq) approach, we found that genes involved in DNA repair, particularly the SOS response program, are upregulated following exposure of the pseudomonad to the DNA-damaging agents mitomycin C and hydrogen peroxide, while genes involved in cell division and primary metabolism are downregulated. The R-tailocin and prophage gene clusters were also upregulated in response to these DNA damaging agents. By combining reverse genetics, transcriptional reporters and chromatin immunoprecipitation sequencing (ChIP-seq), we show that the R-tailocin locus-specific LexA-like regulator PrtR1 represses R-tailocin gene expression by binding directly to the promoter region of the cluster, while the histone-like nucleoid structuring (H-NS) proteins MvaT and MvaV act as master regulators that indirectly regulate R-tailocin cluster expression. Our results suggest that at least these three regulators operate in concert to ensure tight control of R-tailocin expression and cell lytic release in environmental Pseudomonas protegens strains.

|
Scooped by
?
May 12, 11:30 PM
|
The effect of ubiquitous environmental conditions on mutational mechanisms, particularly loss of heterozygosity (LOH) remains poorly understood. Environment induced LOH can rapidly alter the genome and promote disease progression. Using mutation accumulation (MA) lines, we analysed the effect of ubiquitous environmental conditions on mutational mechanisms in a diploid hybrid (S288c/YJM789) baker’s yeast strain. These included blue light, low glucose (calorie restriction), oxidative stress (H2O2), high temperature (37°C), ethanol, and salt (NaCl). The frequency of LOH increased significantly in all environments including calorie restriction relative to the control (YPD). Interestingly, the percentage of the genome covered by LOH varied significantly depending on the condition. For example, the LOH tracts seen in calorie restriction conditions were significantly shorter than those observed in blue light exposure that rapidly homozygotized the genome. We also report a unique mutational signature of blue light exposure comprising LOH, small indels, large deletions and transversion mutations (G:C > T:A; G:C > C:G), with the latter likely to result from the photooxidation of guanine bases. Our results suggest ubiquitous environmental conditions cause LOH but result in distinct mutational signatures due to the type of damage induced and the pathways used to repair them.

|
Scooped by
?
May 12, 11:19 PM
|
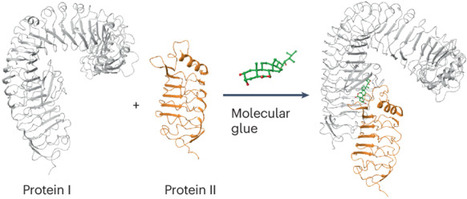
Protein–protein interactions are one of the pillars of all life processes. Many signalling molecules work by promoting and stabilizing these interactions. These molecular ‘glues’ bind simultaneously to two proteins inducing their interaction, which would be otherwise less favorable or non-favorable. Importantly, they can be harnessed for a clinical purpose, but, despite advances in medicine, the wealth of natural molecular glues in plants have only rarely been commercially utilized. These molecular glues may be plant-endogenous or plant-exogenous small molecules or peptides, and they may be involved in many different processes, such as growth promotion or stress response, opening new opportunities for crop protection, along with other applications. In this Review, we analyse the underlying structural motives and molecular interactions in detail, classifying the modes of actions based on their nature (small ligands versus peptides) and receptor classes. We discuss both natural metabolites and mimetics of such compounds, highlighting similarities and differences between signalling pathways and comparing them with relevant mechanisms in mammals. In this Review we discuss the fascinating molecular details of the small molecules and peptides promoting plant protein–protein interactions, and their relevance for plant development and environmental responses.

|
Scooped by
?
May 12, 11:04 PM
|
Host-protective or disease-suppressive microorganisms are emerging as sustainable solutions for controlling crop diseases, such as bacterial wilt. However, the efficacy of biocontrol strategies is often constrained by limited resilience under varying environmental conditions and interactions with native microbial communities in the field. One major challenge is that introduced biocontrol microbes often face suppression by indigenous microbes due to competitive interactions. Synthetic communities (SynCom) offer a promising alternative strategy. However, conventional approaches to assembling SynComs by combining different microbial isolates often result in antagonism and competition among strains, leading to ineffective and inconsistent outcomes. In this study, we assembled a bacterial wilt-suppressive SynCom for tomato, composed of bacterial isolates derived from co-cultured microbial complexes associated with healthy plants. This SynCom demonstrates significant disease-suppressive effects against Ralstonia pseudosolanacearum in tomato seedlings under both axenic and soil conditions. Additionally, our findings suggest the presence of an optimal SynCom colonization level in plants, which is crucial for effective disease suppression. The SynCom also exhibits direct antibiotic activity and modulates the plant-associated microbiome. Our results provide an effective approach to constructing SynComs with consistent and effective disease-suppressive properties within microbial community contexts.

|
Scooped by
?
May 12, 10:09 PM
|
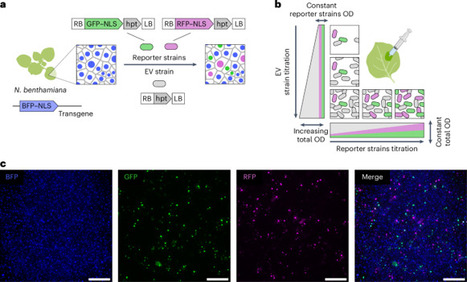
Agrobacterium pathogenesis, which involves transferring T-DNA into plant cells, is the cornerstone of plant genetic engineering. As the applications that rely on Agrobacterium increase in sophistication, it becomes critical to achieve a quantitative and predictive understanding of T-DNA expression at the level of single plant cells. Here we examine if a classic Poisson model of interactions between pathogens and host cells holds true for Agrobacterium infecting Nicotiana benthamiana. Systematically challenging this model revealed antagonistic and synergistic density-dependent interactions between bacteria that do not require quorum sensing. Using various approaches, we studied the molecular basis of these interactions. To overcome the engineering constraints imposed by antagonism, we created a dual binary vector system termed ‘BiBi’, which can improve the efficiency of a reconstituted complex metabolic pathway in a predictive fashion. Our findings illustrate how combining theoretical models with quantitative experiments can reveal new principles of bacterial pathogenesis, impacting both fundamental and applied plant biology. Alamos et al. show that Agrobacterium engages in density-dependent interactions that dictate transformation efficiency and transgene expression. These findings enable a quantitative model to improve metabolic engineering in a predictive manner.

|
Scooped by
?
May 12, 9:30 PM
|
Chromophore-containing proteins (CCPs), including fluorescent and non-fluorescent (chromoproteins), have been widely used in microbiological research. However, several roadblocks often limit their use in non-model bacterial species, including efficient transformation, suitable plasmid origins of replication, and optimal promoter choice. Here, we have engineered a set of 32 shuttle plasmids designed to overcome these roadblocks in an effort to streamline this process for future research. We have selected eight different CCPs: eforCP, YukonOFP, DasherGFP, tinsel Purple, aeBlue, FuGFP, super-folder GFP, and super-folder Cherry2. To broaden the potential host range, we utilized two distinct backbones with p15a either fused to a Francisella origin (FnOri) or to the broad host origin RSF1010 and included a transfer origin (oriT) to facilitate transformation via conjugation. Moreover, we have created versions of each vector, which confer resistance to either kanamycin or chloramphenicol. Lastly, to enable promoter-swapping, we engineered the constitutive pJ23100 promoter element to be flanked by BsaI sites, thereby enabling promoter exchange by the Golden Gate assembly to evaluate CCP expression with different host promoters. To demonstrate the usability of the pKEK-Chrom plasmid series, we evaluated their expression in Escherichia coli, Shewanella oneidensis, and Vibrio alginolyticus. We further demonstrated the utility of promoter swapping in Francisella novicida and validated the functionality of the RSF1010 origin in Acinetobacter baumannii. In summary, the pKEK-Chrom plasmid series provides a palette of different CCPs that streamline their use in non-model gram-negative bacteria.

|
Scooped by
?
May 12, 9:17 PM
|
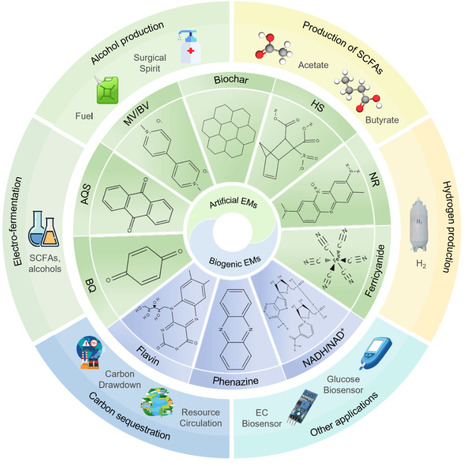
Electron transfer is essential for the production efficiency of value-added products in anaerobic fermentation, such as butanol and ethanol as biofuels, and short-chain fatty acids (SCFAs) including butyric acid and acetic acid as platform chemicals. Electron mediators (EMs), also known as electron shuttles, can facilitate electron transfer to counter irreversible or slow redox reactions that limit fermentation. The addition of EMs has been shown to be an effective strategy to promote fermentation by various bacteria, particularly Clostridium species, for these valuable product syntheses. This paper reviews recent advancements in the application of exogenous electron mediators (EEMs) across various scenarios. Common EEM types, their characteristics, and mechanisms are summarized, and different application scenarios are discussed to elucidate the effect of EEMs. Key technical challenges and future directions for EEM application are also explored.
|

|
Scooped by
?
Today, 1:48 AM
|
Many phenotypic traits, such as fermentation activity, have been shown to be instable due to stochastic gene expression and environmental influence. While previous studies only have obtained understanding at the level of the microbial community, the fate of extraordinary traits of an individual through generations of reproduction has yet to be adequately investigated. This work uses the lactic acid bacteri Lactiplantibacillus plantarum as a research model to study the activity inheritance between parental generations and filial generations. An integrated single-cell manipulation strategy is established, including fluorescent screening using an extracellular pH probe and a microwell array, micropicking using a micropipette, and amplifying an individual bacterium via single-cell culture. Consequently, it is found that daughter bacteria can well inherit the strong acid-producing activity from their parental bacterial individuals, although as the reproduction proceeds over 30 generations, the offspring gradually regresses to the mediocre, thus setting a caveat for the limiting generations for desired inheritance. This is likely due to the deterioration of the cell living environment. This work illustrates the inheritable features of bacterial metabolic traits at the level of individual bacteria and is therefore fundamentally insightful for biotechnological applications like bioenergy production that require consistent or at least predictable metabolic performance.

|
Scooped by
?
Today, 1:35 AM
|
CRISPR-Cas technologies facilitate routine genome engineering of one or a few genes at a time. However, large-scale CRISPR screens with guide RNA libraries remain challenging in plants. Here, we have developed a comprehensive all-in-one CRISPR toolbox for Cas9-based genome editing, cytosine base editing, adenine base editing (ABE), Cas12a-based genome editing and ABE, and CRISPR-Act3.0-based gene activation in both monocot and dicot plants. We evaluated all-in-one T-DNA expression vectors in rice (Oryza sativa, monocot) and tomato (Solanum lycopersicum, dicot) protoplasts, demonstrating their broad and reliable applicability. To showcase the applications of these vectors in CRISPR screens, we constructed guide RNA (gRNA) pools for testing in rice protoplasts, establishing a high-throughput approach to select high-activity gRNAs. Additionally, we demonstrated the efficacy of sgRNA library screening for targeted mutagenesis of ACETOLACTATE SYNTHASE in rice, recovering novel candidate alleles for herbicide resistance. Furthermore, we carried out a CRISPR activation screen in Arabidopsis thaliana, rapidly identifying potent gRNAs for FLOWERING LOCUS T activation that confer an early-flowering phenotype. This toolbox contains 61 versatile all-in-one vectors encompassing nearly all commonly used CRISPR technologies. It will facilitate large-scale genetic screens for loss-of-function or gain-of-function studies, presenting numerous promising applications in plants.

|
Scooped by
?
Today, 1:08 AM
|
Cas9-based genome editing technologies can rapidly generate mutations to probe a diverse array of mutant genotypes. However, aberrant Cas9 nuclease translation and activity can occur despite the use of inducible promoters to control expression, leading to extensive cell death. This background killing caused by promoter leakiness severely limits the application of Cas9 for generating mutant libraries because of the potential for population skew. We demonstrate the utility of temperature sensitive RNA elements as a layer of post-transcriptional regulation to reduce the impact of promoter leak. We observe significant temperature-dependent increases in cell survival when certain RNA thermometers (RNATs) are placed upstream of the cas9 coding sequence. We show that the most highly repressing RNAT, hsp17rep, significantly reduces population skew with a library of characterized guide RNAs (gRNAs). This information should be applicable to all Cas9-based methods and technologies.

|
Scooped by
?
Today, 12:54 AM
|
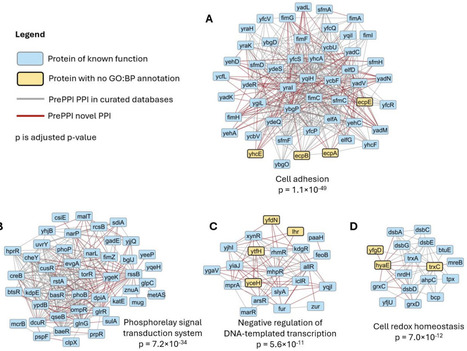
We report on the integration of three methods that are computationally efficient enough to predict, on a proteome-wide scale, whether two proteins are likely to form a binary complex. The methods include PrePPI, which uses three-dimensional structure information as a basis for predictions, Topsy-Turvy which analyzes sequences using a protein language model, and ZEPPI which uses evolutionary information to evaluate protein-protein interfaces. We demonstrate how these methods can be integrated and validate the performance of the integrated method and its separate components at predicting E. coli protein-protein interactions through testing on the HINT high-quality literature-curated database of binary interactions. The integrated method identifies more high-confidence (FPR ≤ 0.001) interactions (~20K) than any of the component methods. The AF3Complex algorithm was used to predict the structures of 400 ppi, and 78% of the integrated method predictions resulted in models deemed accurate by the AF3Complex evaluation score. Notably, essentially all AF3Complex models have at least partially overlapping interfaces with PrePPI models of the complexes. Finally, we clustered the high-confidence E. coli interactome and obtained 385 subnetworks which have high functional coherence defined by enrichment of Gene Ontology Biological Process terms, thus, illustrating that our methods which contain no explicit functional information provide biologically meaningful protein interactions. Biological insights derived from the subnetworks, including the annotation of proteins of unknown function, are discussed in detail. Overall, independent validations support the accuracy of the comprehensive E. coli interactome we have presented.

|
Scooped by
?
Today, 12:09 AM
|
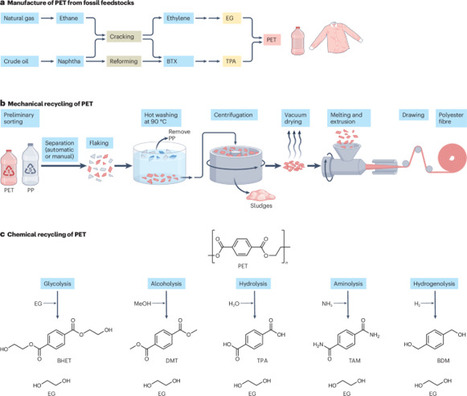
With only a fraction of plastic waste being recovered and the majority incinerated, landfilled or released into the environment, plastic is increasingly accumulating in aquatic and terrestrial ecosystems, leading to widespread pollution. Biocatalysis offers a route to the sustainable recycling of synthetic polyesters, such as polyethylene terephthalate (PET), which is commonly used in textiles, food and beverage packaging, representing a major source of plastic waste. In particular, polyester hydrolases can deconstruct recalcitrant synthetic polymers at scale. This Review discusses the role of biocatalysis in a circular economy of plastics, outlining enzymatic modification and deconstruction strategies for synthetic polyesters. Moreover, protein engineering and computational approaches are examined to design and optimize polyester hydrolases for large-scale recycling in key industrial applications. Finally, the importance of economic viability is highlighted, including strategies to make biocatalysis a cost-effective and sustainable plastics recycling strategy. Plastic pollution could be partly addressed through the biocatalytic recycling of plastic waste streams. This Review discusses the identification and optimization of polyester-degrading enzymes for the large-scale recycling of plastic waste. industry

|
Scooped by
?
May 12, 11:58 PM
|
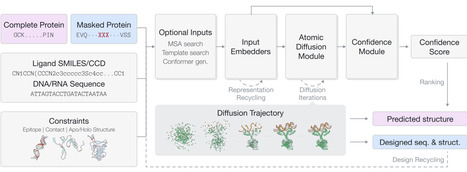
We introduce GeoFlow-V2, a unified atomic diffusion model that seamlessly integrates structure prediction and de novo protein design across multiple biological modalities, including proteins, nucleic acids (DNA/RNA), and small molecules. The model's core innovation lies in its unified architecture that natively handles both structure prediction (when provided with complete input sequences) and generative design (for partially or fully masked inputs) through a shared atomic diffusion process. Through the integration of structure conditioning constraints and the sequence design module, GeoFlow-V2 operates as a fully bidirectional framework, accepting both sequence and structural inputs while generating corresponding outputs. GeoFlow-V2 can also accommodate diverse experimental constraints and prior knowledge, which boosts performance and enables precise control over the folding and design process. Benchmarking against state-of-the-art methods demonstrates GeoFlow-V2's strong performance in both structure prediction and de novo antibody design. We showcase GeoFlow-V2's versatility through protein design cases conditioned on diverse target modalities. To maximize accessibility, we engineer key functionalities of \method and provide a user-friendly web server at https://prot.design for non-commercial research use.

|
Scooped by
?
May 12, 11:33 PM
|
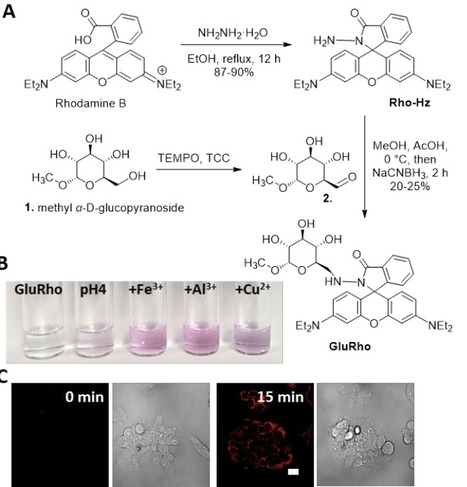
The direct link between sugar uptake and metabolic diseases highlighted the eminent need for molecular tools to detect and evaluate alterations in sugar uptake efficiency as approaches to detect health- and disease-relevant metabolic alterations. However, stringent requirements of the facilitative glucose transporters towards substrate binding and translocation challenge the development of respective fluorescence molecular probes. Based on the state-of-the-art understanding of glucose recognition by facilitative transporters (GLUTs), we designed a glucopyranoside mimic - GluRho - that delivers the “turn-on” rhodamine B to live cells via glucose transport, including major transporters GLUTs 1-4. The accumulation of the GluRho probe is concentration-dependent and is mediated by glucose transported, including major transporters GLUTs 1-4. The high binding affinity achieved through the secondary interaction between the fluorophore and a GLUT protein supports the delivery of the probe in nutrient-rich conditions, facilitating its use as a tool for a direct assessment of glucose GLUT activity in live cells and organisms and across various experimental settings, including uptake evaluation in the presence of sugars, inhibitors, or uptake stimulators. The lack of metabolic contribution to the probe uptake due to the elimination of the phosphorylation site contributes to the high efficacy of the GluRho probe to reflecting alterations in glucose uptake efficiency in live cells, between cell lines, and in multicellular model organisms, such as Drosophila melanogaster. The molecular modeling analysis of GLUT-GluRho complexes with GLUT1 and GLUT2 as two primary transport facilitators provided essential information on GLUT-probe interactions, highlighting the residues facilitating the effective binding and translocation of the probe through trans-ports, thus setting the basis for developing glucose-based glycoconjugates as a cargo-delivering platform.

|
Scooped by
?
May 12, 11:22 PM
|
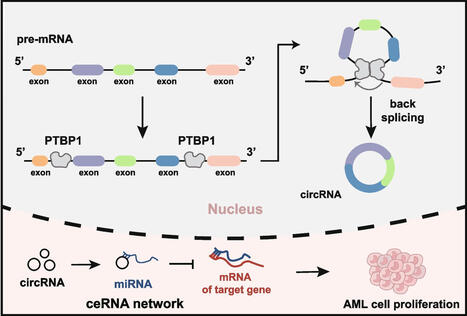
Circular RNAs (circRNAs) are a class of non-coding RNAs generated through back splicing. High expression of circRNAs is often associated with numerous abnormal cellular biological processes. However, the regulatory factors of circRNAs are not fully understood. In this study, we identified PTBP1 as a crucial regulator of circRNA biogenesis through a comprehensive analysis of the whole transcriptome profiles across 10 diverse cell lines. Knockdown of PTBP1 led to a significant decrease in circRNA expression, concomitant with a distinct reduction in cell proliferation. To investigate the regulatory mechanism of PTBP1 on circRNA biogenesis, we constructed a minigene reporter based on SPPL3 gene. The results showed that PTBP1 can bind to the flanking introns of circSPPL3, and the mutation of PTBP1 motif impedes the back splicing of circSPPL3. Subsequently, to demonstrate that this observation is not an exception, the comprehensive regulatory effects of PTBP1 on circRNAs were confirmed by miniGFP, reflecting the necessity of the binding site in the flanking introns. Analysis of data from clinical samples showed that both PTBP1 and circRNAs exhibited substantial upregulation in acute myeloid leukemia, further demonstrating a potential role for PTBP1 in promoting circRNA biogenesis under in vivo conditions. Competitive endogenous RNA (ceRNA) network revealed that PTBP1-associated circRNAs participated in biological processes associated with cell proliferation. In summary, our study is the first to identify the regulatory effect of PTBP1 on circRNA biogenesis and indicates a possible link between PTBP1 and circRNA expression in leukemia.

|
Scooped by
?
May 12, 11:15 PM
|
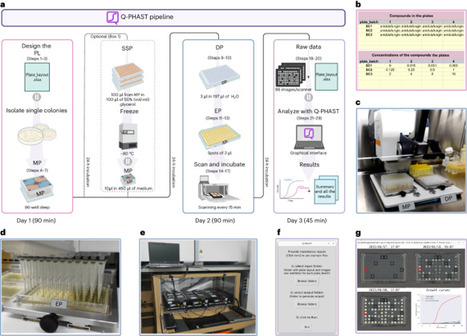
The characterization of antimicrobial susceptibility and other relevant phenotypes in large collections of microbial isolates is a common need across research and clinical microbiology laboratories. Robotization provides unprecedented throughput but involves costs that are prohibitive for the average laboratory. Here, using affordable materials and open-source software, we developed Q-PHAST (Quantitative PHenotyping and Antimicrobial Susceptibility Testing), a unique solution for cost-effective, large-scale phenotyping in a standard microbiology laboratory. Single colonies are grown in a deep 96-well master plate, from which diluted aliquots are used to generate 96 spots on different experimental plates containing solid medium with the substance and concentration of interest. These plates are incubated on inexpensive flatbed scanners that monitor the growth of each spot by obtaining images every 15 min. A simple, python-based software, which can be used via a graphical interface on various operating systems ( https://github.com/Gabaldonlab/Q-PHAST ), analyzes the images to infer growth, fitness (e.g., doubling rate) and susceptibility (e.g., minimum inhibitory concentration) measures. With <120 min of hands-on time per day for three consecutive days, ready-to-use results are obtained and presented in tables or graphs. This solution enables non-experts with limited resources to perform accurate quantitative phenotyping on hundreds of strains in parallel. This Q-PHAST (Quantitative PHenotyping and Antimicrobial Susceptibility Testing) protocol provides a unique solution for cost-effective, large-scale phenotyping in a standard microbiology laboratory using affordable materials and open-source software.

|
Scooped by
?
May 12, 10:11 PM
|
Results from a phase III clinical trial suggest that taking ubrogepant at the first sign of an oncoming migraine can prevent pre-headache fatigue and light sensitivity.

|
Scooped by
?
May 12, 9:49 PM
|
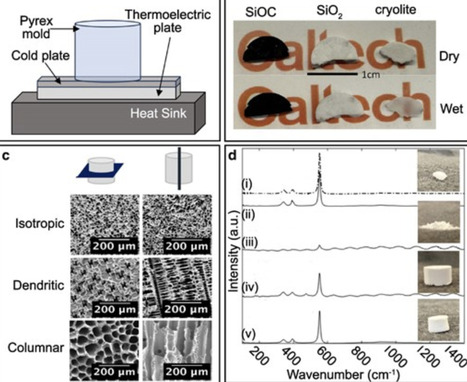
Microbes inhabiting complex porous microenvironments in sediments and aquifers catalyze reactions that are critical to global biogeochemical cycles and ecosystem health. However, the opacity and complexity of porous sediment and rock matrices have considerably hindered the study of microbial processes occurring within these habitats. Here, we generated microbially compatible, optically transparent mineral scaffolds to visualize and investigate microbial colonization and activities occurring in these environments, in laboratory settings and in situ. Using inexpensive synthetic cryolite mineral, we produced optically transparent scaffolds mimicking the complex 3D structure of sediments and rocks by adapting a suspension-based, freeze-casting technique commonly used in materials science. Fine-tuning of parameters, such as freezing rate and choice of solvent, provided full control of pore size and architecture. The combined effects of scaffold porosity and structure on the movement of microbe-sized particles, tested using velocity tracking of fluorescent beads, showed diverse yet reproducible behaviors. The scaffolds we produced are compatible with epifluorescence microscopy, allowing the fluorescence-based identification of colonizing microbes by DNA-based staining and fluorescence in situ hybridization (FISH) to depths of 100 µm. Additionally, Raman spectroscopy analysis indicates minimal background signal in regions used for measuring deuterium and 13C enrichment in microorganisms, highlighting the potential to directly couple D2O or 13C stable isotope probing and Raman-FISH for quantifying microbial activity at the single-cell level. To demonstrate the relevance of cryolite scaffolds for environmental field studies, we visualized their colonization by diverse microorganisms within rhizosphere sediments of a coastal seagrass plant using epifluorescence microscopy. The tool presented here enables highly resolved, spatially explicit, and multimodal investigations into the distribution, activities, and interactions of underground microbes typically obscured within opaque geological materials until now.

|
Scooped by
?
May 12, 9:24 PM
|
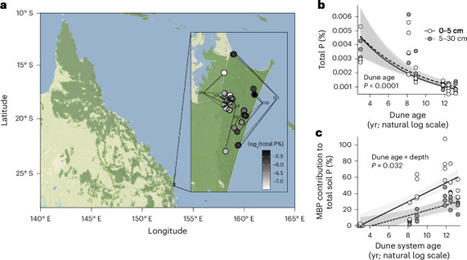
Long-term terrestrial ecosystem development is characterized by declining soil phosphorus (P) and a corresponding increase in biological P limitation. The function of P-limited ecosystems relies on efficient use of P by soil microorganisms, but the physiological strategies used by microorganisms to manage P scarcity during ecosystem development are unknown. Here, by applying recent advances in soil metabolomic techniques to samples collected from a ~700,000-year chronosequence of ecosystem development in eastern Australia, we show that soil microbial physiological strategies for P efficiency include a high proportion of non-phosphorous membrane lipids along with substantial intracellular carbon storage. These strategies—which proliferate during primary succession and are maximized in retrogressive, P-depleted ecosystems—uphold microbial carbon limitation, triple modelled P-mineralization potential and can conserve close to double the P contained in the aboveground biomass of vegetation. These findings transform our understanding of terrestrial ecosystems by revealing a strong yet overlooked interplay between the ecophysiology of soil microorganisms and the long-term trajectory of ecosystem development. Microbial physiology conserves phosphorus by reducing phosphorus contributions in membrane lipids and increasing intracellular carbon storage during long-term ecosystem development, according to measurements of a ~700,000–year chronosequence across an Australian dune system.
|
 Your new post is loading...
Your new post is loading...





















the fs8.1 mutation allele is present in modern processing lines but largely absent in fresh-market varieties.