Your new post is loading...
 Your new post is loading...

|
Scooped by
Kamoun Lab @ TSL
June 26, 2012 9:39 AM
|
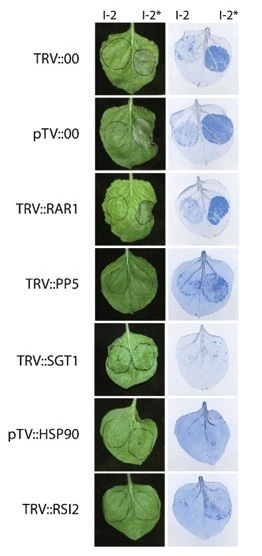
Race-specific disease resistance in plants depends on the presence of resistance (R) genes. Most R genes encode NB-ARC-LRR proteins that carry a C-terminal leucine-rich repeat (LRR). Of the few proteins found to interact with the LRR domain, most have proposed (co)chaperone activity. Here, we report the identification of RSI2 (Required for Stability of I-2) as a protein that interacts with the LRR domain of the tomato R protein I-2. RSI2 belongs to the family of small heat shock proteins (sHSPs or HSP20s). HSP20s are ATP-independent chaperones that form oligomeric complexes with client proteins to prevent unfolding and subsequent aggregation. Silencing of RSI2-related HSP20s in Nicotiana benthamiana compromised the hypersensitive response that is normally induced by auto-active variants of I-2 and Mi-1, a second tomato R protein. As many HSP20s have chaperone properties, the involvement of RSI2 and other R protein (co)chaperones in I-2 and Mi-1 protein stability was examined. RSI2 silencing compromised the accumulation of full-length I-2 in planta, but did not affect Mi-1 levels. Silencing of heat shock protein 90 (HSP90) and SGT1 led to an almost complete loss of full-length I-2 accumulation and a reduction in Mi-1 protein levels. In contrast to SGT1 and HSP90, RSI2 silencing led to accumulation of I-2 breakdown products. This difference suggests that RSI2 and HSP90/SGT1 chaperone the I-2 protein using different molecular mechanisms. We conclude that I-2 protein function requires RSI2, either through direct interaction with, and stabilization of I-2 protein or by affecting signalling components involved in initiation of the hypersensitive response.

|
Scooped by
Kamoun Lab @ TSL
June 25, 2012 10:28 AM
|
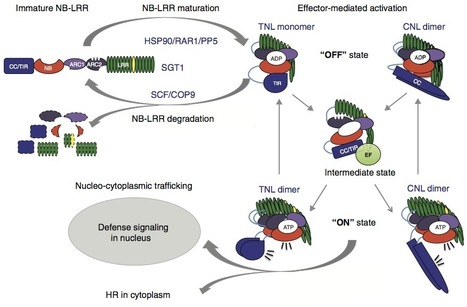
Many plant disease resistance (R) proteins belong to the family of nucleotide-binding-leucine rich repeat (NB-LRR) proteins. NB-LRRs mediate recognition of pathogen-derived effector molecules and subsequently activate host defence. Their multi-domain structure allows these pathogen detectors to simultaneously act as sensor, switch and response factor. Structure–function analyses and the recent elucidation of the 3D structures of subdomains have provided new insight in how these different functions are combined and what the contribution is of the individual subdomains. Besides interdomain contacts, interactions with chaperones, the proteasome and effector baits are required to keep NB-LRRs in a signalling-competent, yet auto-inhibited state. In this review we explore operational models of NB-LRR functioning based on recent advances in understanding their structure. ► The N-terminal CC and TIR domain of Mla1 and L6 form homo dimers that are required and sufficient to induce cell death. ► Steady-state levels of signalling-competent NB-LRR R proteins are co-regulated by chaperones and the proteasome. ► NB-LRR activation is a multistep process requiring fine-tuned intramolecular interactions between co-evolved subdomains. ► 3D modelling of NB-LRR structures aids predicting the conformational changes underlying R protein function and activity. ► Changes in the conformational fold of NB-LRR R proteins correlate with distinct subcellular localisations.

|
Scooped by
Kamoun Lab @ TSL
June 25, 2012 2:30 AM
|
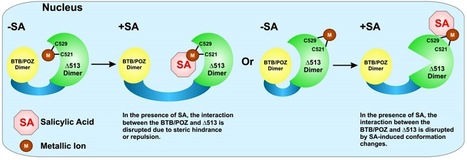
> NPR1 is a salicylic acid (SA) receptor, binding specifically to SA via Cys521/529
> NPR1 binds copper in vivo via Cys521/529, and metals are required for SA binding
> SA directly regulates the conformation of NPR1 by deoligomerizing NPR1 into a dimer
> The NPR1 BTB/POZ domain autoinhibits the function of the NPR1 transactivation domain Salicylic acid (SA) is an essential hormone in plant immunity, but its receptor has remained elusive for decades. The transcriptional coregulator NPR1 is central to the activation of SA-dependent defense genes, and we previously found that Cys521 and Cys529 of Arabidopsis NPR1's transactivation domain are critical for coactivator function. Here, we demonstrate that NPR1 directly binds SA, but not inactive structural analogs, with an affinity similar to that of other hormone-receptor interactions and consistent with in vivo Arabidopsis SA concentrations. Binding of SA occurs through Cys521/529 via the transition metal copper. Mechanistically, our results suggest that binding of SA causes a conformational change in NPR1 that is accompanied by the release of the C-terminal transactivation domain from the N-terminal autoinhibitory BTB/POZ domain. While NPR1 is already known as a link between the SA signaling molecule and defense-gene activation, we now show that NPR1 is the receptor for SA.

|
Scooped by
Kamoun Lab @ TSL
June 24, 2012 3:56 AM
|
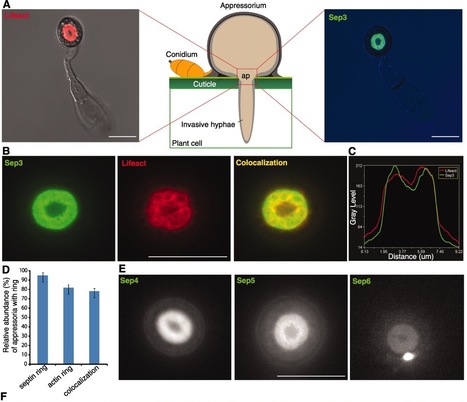
To cause rice blast disease, the fungus Magnaporthe oryzae develops a pressurized dome-shaped cell called an appressorium, which physically ruptures the leaf cuticle to gain entry to plant tissue. Here, we report that a toroidal F-actin network assembles in the appressorium by means of four septin guanosine triphosphatases, which polymerize into a dynamic, hetero-oligomeric ring. Septins scaffold F-actin, via the ezrin-radixin-moesin protein Tea1, and phosphatidylinositide interactions at the appressorium plasma membrane. The septin ring assembles in a Cdc42- and Chm1-dependent manner and forms a diffusion barrier to localize the inverse-bin-amphiphysin-RVS–domain protein Rvs167 and the Wiskott-Aldrich syndrome protein Las17 at the point of penetration. Septins thereby provide the cortical rigidity and membrane curvature necessary for protrusion of a rigid penetration peg to breach the leaf surface.

|
Scooped by
Kamoun Lab @ TSL
June 23, 2012 6:58 AM
|
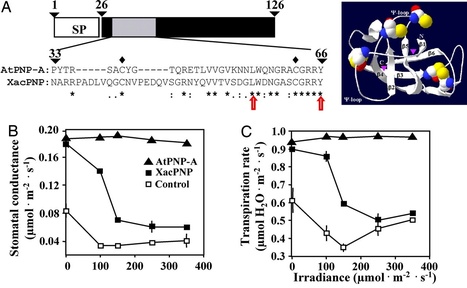
Plant natriuretic peptides (PNPs) are a class of extracellular, systemically mobile molecules that elicit a number of plant responses important in homeostasis and growth. The bacterial citrus pathogen, Xanthomonas axonopodis pv. citri, also contains a gene encoding a PNP-like protein, XacPNP, that shares significant sequence similarity and identical domain organization with plant PNPs but has no homologues in other bacteria. We have expressed and purified XacPNP and demonstrated that the bacterial protein alters physiological responses including stomatal opening in plants. Although XacPNP is not expressed under standard nutrient rich culture conditions, it is strongly induced under conditions that mimic the nutrient poor intercellular apoplastic environment of leaves, as well as in infected tissue, suggesting that XacPNP transcription can respond to the host environment. To characterize the role of XacPNP during bacterial infection, we constructed a XacPNP deletion mutant. The lesions caused by this mutant were more necrotic than those observed with the wild-type, and bacterial cell death occurred earlier in the mutant. Moreover, when we expressed XacPNP in Xanthomonas axonopodis pv. vesicatoria, the transgenic bacteria caused less necrotic lesions in the host than the wild-type. In conclusion, we present evidence that a plant-like bacterial PNP can enable a plant pathogen to modify host responses to create conditions favorable to its own survival.

|
Scooped by
Kamoun Lab @ TSL
June 20, 2012 3:40 PM
|
Paleontological J: The most ancient terrestrial lichen Winfrenatia reticulata : A new find and new interpretation (2009)
http://www.springerlink.com/content/g8l8708r5gr36646/ Silicified fossils from Rhynie cherts in Scotland are studied. A lichen belonging to the genus Winfrenatia is detected and studied. This oldest terrestrial lichen is dated to the Pragian (=Siegenian) of the Early Devonian (407 to 411 million years ago). New characters of the lichen are described, and their new interpretation is given. The main component of the lichen thallus is a filamentous cyanobacterium (Nostocales). Structures which were interpreted as fungal hyphae are probably hollow sheaths of this cyanobacterium. Mycobiont hyphae develop at the base of the thallus and symbiose with a coccoid cyanobacterium. Thus, Winfrenatia reticulata is a three-parted organism, constituted of a mycobiont and filamentous and coccoid cyanobacteria.

|
Scooped by
Kamoun Lab @ TSL
June 20, 2012 11:46 AM
|
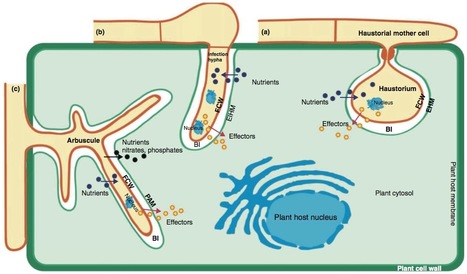
Both mutualistic and biotrophic pathogenic fungi rely on living host plants for growth and reproduction and must modify host cell structure and function for successful infection. The deployment of a diverse set of secreted virulence determinants referred to as ‘effectors’, many of which are directly delivered into the host cell, is postulated to be the key to host infection. This review provides a snapshot of the current progress in fungal effector biology. Recent genome sequencing of rust and powdery mildew obligate biotrophs has provided insight into the repertoires of potential effectors of these highly specialised pathogens. Identification of the first host-translocated effectors from mutualistic fungi has revealed that these fungi also manipulate host cells through effectors. The biological activities of some fungal effectors are just beginning to be revealed, while much uncertainty still surrounds the mechanisms of transport into host cells.

|
Rescooped by
Kamoun Lab @ TSL
from biotech-translations
June 19, 2012 12:24 PM
|
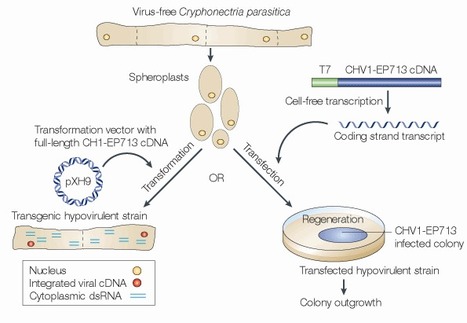
Whereas most mycoviruses lead 'secret lives', some reduce the ability of their fungal hosts to cause disease in plants. This property, known as hypovirulence, has attracted attention owing to the importance of fungal diseases in agriculture and the limited strategies that are available for the control of these diseases. Using one pathogen to control another is appealing, both intellectually and ecologically. The recent development of an infectious cDNA-based reverse genetics system for members of the Hypoviridae mycovirus family has enabled the analysis of basic aspects of this fascinating virus–fungus–plant interaction, including virus–host interactions, the mechanisms underlying fungal pathogenesis, fungal signalling pathways and the evolution of RNA silencing. Such systems also provide a means for engineering mycoviruses for enhanced biocontrol potential.
Via Plant-Microbe Interaction

|
Scooped by
Kamoun Lab @ TSL
June 19, 2012 11:58 AM
|
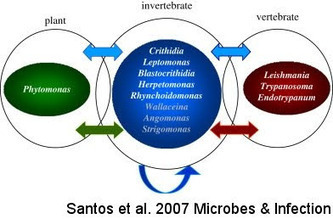
While I was trying to come up with something quick to blog about, got a couple updates in Google Reader from J. Eukaryotic Microbiol, among them a paper on... trypanosomatids living in coconut tree phloem! Somehow, you don't typically think of plants being invaded by motile, flagellate things, but on a second thought: why not? The phloem is a vessel, and while perhaps there's no need to run away from macrophages or anything, there's no particular harm in retaining the ability to swim around. Especially if your other life happens in...insects!

|
Scooped by
Kamoun Lab @ TSL
June 19, 2012 8:15 AM
|
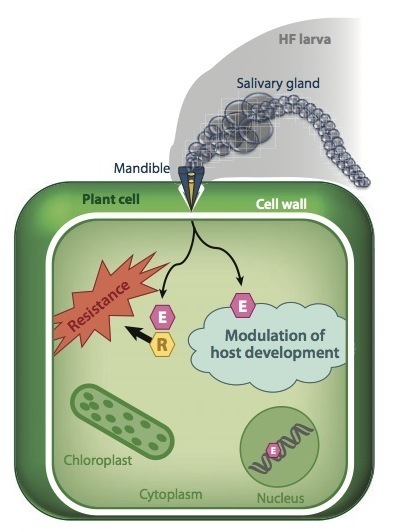
Gall midges constitute an important group of plant-parasitic insects. The Hessian fly (HF; Mayetiola destructor), the most investigated gall midge, was the first insect hypothesized to have a gene-for-gene interaction with its host plant, wheat (Triticum spp.). Recent investigations support that hypothesis. The minute larval mandibles appear to act in a manner that is analogous to nematode stylets and the haustoria of filamentous plant pathogens. Putative effector proteins are encoded by hundreds of genes and expressed in the HF larval salivary gland. Cultivar-specific resistance (R) genes mediate a highly localized plant reaction that prevents the survival of avirulent HF larvae. Fine-scale mapping of HF avirulence (Avr) genes provides further evidence of effector-triggered immunity (ETI) against HF in wheat. Taken together, these discoveries suggest that the HF, and other gall midges, may be considered biotrophic, or hemibiotrophic, plant pathogens, and they demonstrate the potential that the wheat-HF interaction has in the study of insect-induced plant gall formation.

|
Scooped by
Kamoun Lab @ TSL
June 18, 2012 5:50 AM
|
Tweets from the The Sainsbury Laboratory EMBO Practical Course on Plant Microbe Interactions...

|
Scooped by
Kamoun Lab @ TSL
June 16, 2012 12:41 PM
|
New Zealand - Havelock North Plant & Food Research scientist Dr Ian Horner is using a technique developed for orchards to attempt to control an organism devastating kauri forests. "It is a very significant disease," he said. "It will kill kauri trees - it doesn't muck around. It seems to be a wide range of situations that kauri are being diseased and killed." The newly discovered Phytophthora taxon Agathis (PTA) was first discovered in 1ha of a partially logged kauri forest on Great Barrier Island in the 1970s. "My understanding is there is now 30ha affected on the island. "It was initially misnamed and seen as a fairly minor problem. As it turns out, it is the same organism we are now finding in kauri forests in many parts of the Northland and Auckland regions."

|
Scooped by
Kamoun Lab @ TSL
June 15, 2012 4:39 AM
|
Recent years have seen an explosion of new high throughput technologies which in turn have enabled ground-breaking discoveries in plant sciences. Next generation sequencing and advances in genome assembly and analysis are helping to crack the code of even the most complex genomes. Transcriptomics, proteomics and metabolomics are leading the way in providing new insights into the inner workings of plant cells and plant cell-cell communications. Equally importantly, the cell biology toolbox – previously mainly restricted to animal and yeast cells – has finally been built up in plants allowing researchers to explore the fundamental questions in plant cell biology and compare plant solutions with those of other organisms. Importantly, the ‘omics’ and cell biology tools also shed light on plant-environment interactions, including responses to abiotic signals and plant-pathogen interactions. This conference will focus on the recent breakthroughs in plant biology, enabled by the latest methodological developments.
|

|
Scooped by
Kamoun Lab @ TSL
June 25, 2012 10:34 AM
|
Plant resistance (R) proteins mediate race-specific immunity and initiate host defenses that are often accompanied by a localized cell-death response. Most R proteins belong to the NB-LRR protein family as they carry a central NB-ARC domain fused to an LRR domain. The CC domain at the N-terminus of some Solanaceous NB-LRR proteins is extended with a solanaceae domain (SD). Tomato Mi-1.2, which confers resistance against nematodes, white flies, psyllids and aphids, encodes a typical SD-CNL protein. Here, we analyzed the role of the extended N-terminus for Mi-1.2 activation. Removal of the first part of the N-terminus (Nt1) induced Mi-1.2-mediated cell death that could be suppressed by over-expression of the second half of the N-terminal region (Nt2). Yet, autoactivating NB-ARC-LRR mutants require in trans co-expression of the N-terminal region to induce cell death, indicating that the N-terminus functions both as a negative and a positive regulator. Based on secondary structure predictions we could link both functions to three distinct subdomains; a typical CC domain and two novel, structurally-conserved helical subdomains called SD1 and SD2. A negative regulatory function could be assigned to the SD1 whereas SD2 and the CC together function as positive regulators of Mi-1.2 mediated cell death.

|
Rescooped by
Kamoun Lab @ TSL
from immunity
June 25, 2012 7:56 AM
|
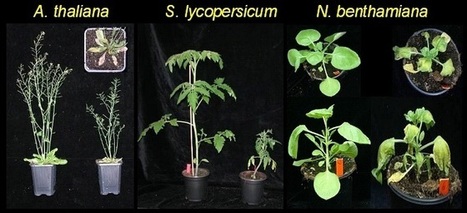
Verticillium spp. are soil-borne plant pathogens that cause vascular wilt diseases in many economically important crops. Verticillium wilt diseases are difficult to control due to the long viability of the resting structures, the broad host range of the pathogens, and the inability of fungicides to eliminate the pathogens once they have entered the xylem tissues of the plant. In our research we employ tomato (Solanum lycopersicum), Australian tobacco (Nicotiana benthamiana) and thale cress (Arabidopsis thaliana) as model plants to investigate the biology of Verticillium wilt diseases. Here is a Verticillium pathogen profile for more information on the pathogen http://www.php.wur.nl/NR/rdonlyres/0C39DFED-E598-43CD-9490-482FEAB6F582/70434/2006FradinMPP.pdf
Via Plant-Microbe Interaction

|
Scooped by
Kamoun Lab @ TSL
June 24, 2012 4:10 AM
|
There’s a microscopic fungus that can starve nations and punch through Kevlar. It kills on such as scale that its effects can be seen from space. It’s called Magnaporthe oryzae and it causes a disease known as rice blast. The fungus doesn’t infect humans, but it does kill rice. It kills a lot of rice, destroying up to 30 per cent of the world’s total crop every year – enough to feed 60 million people. Slowly, scientists have worked out how this cereal killer claims its victims. Nick Talbot from the University of Exeter has spent decades piecing together intricate molecular jig that allows the fungus to create its dome, build its pressure, and enter its host. He was, for example, the one who discovered the role of glycerol, back in 1997. Now, he has solved another mystery: how the appressorium channels its pressure down into its host...

|
Scooped by
Kamoun Lab @ TSL
June 23, 2012 7:12 AM
|
Xanthomonas XacPNP is an effector protein that is similar to Ave1 produced by the plant pathogenic fungus Verticillium dahliae and other fungi. Ave1 is recognized by the surface receptor-like protein Ve1 of tomato. See http://bit.ly/yjdSlf and http://bit.ly/Nkyzb8 The XacPNP paper did not provide the GenBank accession, so here it is: >gi|21243383
MGIVMKHKILLGFSVAAIGLLFSSAAFADIGTISFYGNNARRPADLVQGCNVPEDQVSGRNYQVVTVSDGLWDNGASCGRRYRMRCISTPVKHSCTASTIDVIVVGRCPNGRCTVGGRDVTMKIAFNRYSLLVQARTAPWANI

|
Rescooped by
Kamoun Lab @ TSL
from cell biology
June 21, 2012 12:08 PM
|
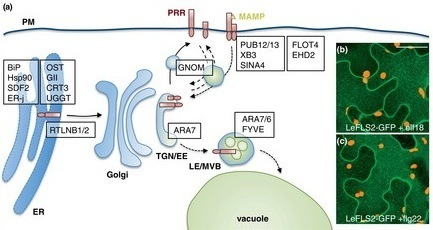
FLS2: a model for studying subcellular reprogramming

|
Rescooped by
Kamoun Lab @ TSL
from effectors
June 20, 2012 2:52 PM
|
Tree rusts are fungal diseases causing dusty orange or brown spots (pustules) on the leaves of poplar, willow, birch, plum and five-needled pine. Tree rusts cause eye-catching infections on the leaves of some trees, particularly Populus spp. (poplar), Salix spp. (willow), Betula spp. (birch), plum and Pinus spp. (five-needled pines). Tree rusts may be seen from spring until autumn for trees that lose their leaves in winter (deciduous) and all year on evergreens. These are the rust fungi involved: > Poplar rust is caused by several species of Melampsora > Willow rust is also caused by several species of Melampsora, but not the same as those infecting poplars > Birch rust is caused by Melampsoridium betulinumPlum rust is caused by Tranzschelia pruni-spinosae var. discolor > Five-needled pines are infected by white pine blister rust, caused by Cronartium ribicola
Via Plant-Microbe Interaction

|
Rescooped by
Kamoun Lab @ TSL
from cell biology
June 20, 2012 2:57 AM
|
Current Opinion Plant Biol: The INs and OUTs of pattern recognition receptors at the cell surface (2012)
http://dx.doi.org/10.1016/j.pbi.2012.05.004 Pattern recognition receptors (PRRs) enable plants to sense non-self molecules displayed by microbes to mount proper defense responses or establish symbiosis. In recent years the importance of PRR subcellular trafficking to plant immunity has become apparent. PRRs traffic through the endoplasmatic reticulum (ER) and the Golgi apparatus to the plasma membrane, where they recognize their cognate ligands. At the plasma membrane, PRRs can be recycled or internalized via endocytic pathways. By using genetic and biochemical tools in combination with bioimaging, the trafficking pathways and their role in PRR perception of microbial molecules are now being revealed. Highlights: ► PRRs are present at the plasma membrane and this enables MAMP recognition. ► Subcellular localization of several PRRs have been revealed in the recent years. ► Insights into PRR trafficking pathways and potential regulators together with the interception of the PRR outputs are being obtained.
Via Plant-Microbe Interaction

|
Scooped by
Kamoun Lab @ TSL
June 19, 2012 12:12 PM
|
Parasitic biotrophic interactions are molecularly based on a battery of effector-molecules which are used by the invader to manipulate the host without killing it. Effectormolecules might be used to camouflage the pathogen against recognition, might suppress host-defense responses by detoxifying or sequestering harmful plant compounds or might redirect host developmental processes or manipulate the host metabolism for the reproductive success of the pathogen. An effector in the context of biotrophic interactions can be any molecule that is released by the biotroph and interferes directly or indirectly with the host cell and thereby affects the interaction between the two organisms. Thus, effectors can be secreted proteins but also non-proteinous secreted molecules. Effectors can promote infection but can also be specifically recognized and trigger defense responses of the host. Thus, the term "effector" is neutral and can imply a positive or negative impact on virulence.

|
Scooped by
Kamoun Lab @ TSL
June 19, 2012 9:51 AM
|
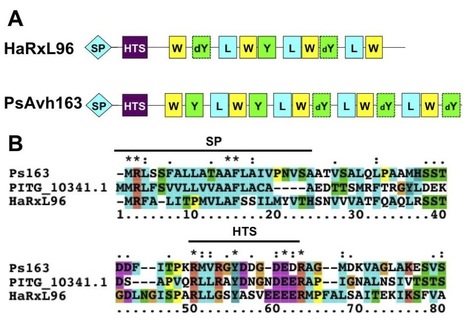
Diverse pathogens secrete effector proteins into plant cells to manipulate host cellular processes. Oomycete pathogens contain large complements of predicted effector genes defined by an RXLR host cell entry motif. The genome of Hyaloperonospora arabidopsidis (Hpa, downy mildew of Arabidopsis) contains at least 134 candidate RXLR effector genes. Only a small subset of these genes is conserved in related oomycetes from the Phytophthora genus. Here, we describe a comparative functional characterization of the Hpa RXLR effector gene HaRxL96 and a homologous gene, PsAvh163, from the soybean pathogen Phytophthora sojae. HaRxL96 and PsAvh163 are induced during early stages of infection and carry a functional RXLR motif that is sufficient for protein uptake into plant cells. Both effectors can suppress immune responses in soybean. HaRxL96 suppresses immunity in Nicotiana benthamiana, while PsAvh163 induces an HR-like cell death response in Nicotiana that is dependent on RAR1 and Hsp90.1. Transgenic Arabidopsis plants expressing HaRxL96 or PsAvh163 exhibit elevated susceptibility to virulent and avirulent Hpa as well as decreased callose deposition in response to non-pathogenic P. syringae. Both effectors interfere with defense marker gene induction, but do not affect salicylic acid biosynthesis. Together, these experiments demonstrate that evolutionarily conserved effectors from different oomycete species can suppress immunity in plant species that are divergent from the source pathogen’s host.

|
Scooped by
Kamoun Lab @ TSL
June 18, 2012 3:55 PM
|
The particular rust tormenting your lavatera is Puccinia malvacearum. It can also attack several other members of this plant family, including other lavatera species, hollyhock, abutilon (flowering maple), hibiscus and weedy mallows, including the one we sometimes call "cheeseweed." Since I looked at your plant, I found two more tree mallows with the same disease, in different parts of San Francisco. Because of this, I encourage readers to check their lavatera, hollyhocks, abutilon and hibiscus for symptoms. Weedy mallows are often infected with this rust while they are still quite small, so pull any you see in your garden to prevent them from spreading the disease. They have dark green, rounded leaves with toothed edges, and, when mature, small pink, lavatera-like blossoms.

|
Scooped by
Kamoun Lab @ TSL
June 17, 2012 9:25 AM
|
XopN is a type III effector protein from Xanthomonas campestris pathovar vesicatoria that suppresses PAMP-triggered immunity (PTI) in tomato. Previous work reported that XopN interacts with the tomato 14-3-3 isoform TFT1; however, TFT1's role in PTI and/or XopN virulence was not determined. Here we show that TFT1 functions in PTI and is a XopN virulence target. Virus-induced gene silencing of TFT1 mRNA in tomato leaves resulted in increased growth of Xcv ΔxopN and Xcv ΔhrpF demonstrating that TFT1 is required to inhibit Xcv multiplication. TFT1 expression was required for Xcv-induced accumulation of PTI5, GRAS4, WRKY28, and LRR22 mRNAs, four PTI marker genes in tomato. Deletion analysis revealed that the XopN C-terminal domain (amino acids 344–733) is sufficient to bind TFT1. Removal of amino acids 605–733 disrupts XopN binding to TFT1 in plant extracts and inhibits XopN-dependent virulence in tomato, demonstrating that these residues are necessary for the XopN/TFT1 interaction. Phos-tag gel analysis and mass spectrometry showed that XopN is phosphorylated in plant extracts at serine 688 in a putative 14-3-3 recognition motif. Mutation of S688 reduced XopN's phosphorylation state but was not sufficient to inhibit binding to TFT1 or reduce XopN virulence. Mutation of S688 and two leucines (L64,L65) in XopN, however, eliminated XopN binding to TFT1 in plant extracts and XopN virulence. L64 and L65 are required for XopN to bind TARK1, a tomato atypical receptor kinase required for PTI. This suggested that TFT1 binding to XopN's C-terminal domain might be stabilized via TARK1/XopN interaction. Pull-down and BiFC analyses show that XopN promotes TARK1/TFT1 complex formation in vitro and in planta by functioning as a molecular scaffold. This is the first report showing that a type III effector targets a host 14-3-3 involved in PTI to promote bacterial pathogenesis.

|
Scooped by
Kamoun Lab @ TSL
June 15, 2012 6:09 AM
|
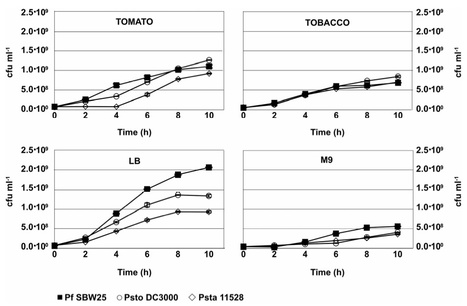
The plant apoplast is the intercellular space that surrounds plant cells, in which metabolic and physiological processes relating to cell wall biosynthesis, nutrient transport, and stress responses occur. The apoplast is also the primary site of infection for hemibiotrophic pathogens such as P. syringae, which obtain nutrients directly from apoplastic fluid. We have used apoplastic fluid extracted from healthy tomato leaves as a growth medium for Pseudomonas spp. in order to investigate the role of apoplastic nutrients in plant colonization by Pseudomonas syringae. We have confirmed that apoplast extracts mimic some of the environmental and nutritional conditions that bacteria encounter during apoplast colonization by demonstrating that expression of the plant-induced type III protein secretion pathway is upregulated during bacterial growth in apoplast extracts. We used a modified phenoarray technique to show that apoplast-adapted P. syringae pv. tomato DC3000 expresses nutrient utilization pathways that allow it to use sugars, organic acids, and amino acids that are highly abundant in the tomato apoplast. Comparative analyses of the nutrient utilization profiles of the genome-sequenced strains P. syringae pv. tomato DC3000, P. syringae pv. syringae B728a, P. syringae pv. phaseolicola 1448A, and the unsequenced strain P. syringae pv. tabaci 11528 with nine other genome-sequenced strains of Pseudomonas provide further evidence that P. syringae strains are adapted to use nutrients that are abundant in the leaf apoplast. Interestingly, P. syringae pv. phaseolicola 1448A lacks many of the nutrient utilization abilities that are present in three other P. syringae strains tested, which can be directly linked to differences in the P. syringae pv. phaseolicola 1448A genome.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...