Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
December 1, 2023 8:46 AM
|
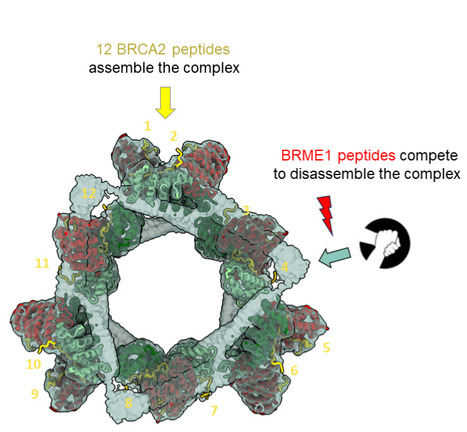
The large ring-shaped BRCA2-HSF2BP complex is disassembled by BRME1, thus promoting Homologous Recombination: a cryo-EM study.
A short disordered and conserved motif of the tumor suppressor BRCA2 triggers the assembly of a large ring-shaped 24-mer of the meiotic HSF2BP protein, whereas the disordered C-terminal motif of the meiotic BRME1 dissociates this ring complex. In meiotic homologous recombination (HR), BRCA2 facilitates loading of the recombinases RAD51 and DMC1 at the sites of double-strand breaks (DSB). The HSF2BP-BRME1 complex interacts with BRCA2. Its absence causes a severe reduction in recombinase loading at meiotic DSB. We previously showed that, in somatic cancer cells ectopically producing HSF2BP, DNA damage can trigger HSF2BP-dependent degradation of BRCA2, which prevents HR. Here we report that, upon binding to BRCA2, HSF2BP forms octameric rings that are able to assemble into a large ring-shaped 24-mer. Addition of BRME1 leads to dissociation of both of these ring structures, and cancels the disruptive effect of HSF2BP on cancer cell resistance to DNA damage. It also prevents BRCA2 degradation during inter-strand DNA crosslink repair in Xenopus egg extracts. We propose that the control of HSF2BP-BRCA2 oligomerization by BRME1 ensures timely assembly of the ring complex that concentrates BRCA2 and controls its turnover, thus promoting HR. More information: https://www.science.org/doi/10.1126/sciadv.adi7352 Contact: Sophie ZINN JUSTIN <sophie.zinn@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 23, 2023 5:08 AM
|
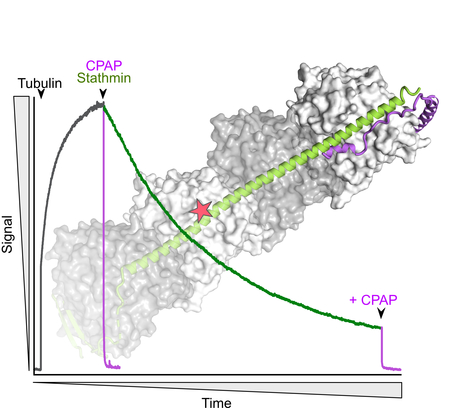
The C-terminus of stathmin-like proteins governs the stability of their complexes with tubulin
Identification of a novel mechanism for the regulation of the stathmin:tubulin interaction. Microtubule dynamics is modulated by many cellular factors including stathmin family proteins. Vertebrate stathmins sequester two ab-tubulin heterodimers into a tight complex that cannot be incorporated in microtubules. Stathmins are regulated at the expression level during development and among tissues; they are also regulated by phosphorylation. We have studied the dissociation kinetics of tubulin:stathmin assemblies in presence of different tubulin-binding proteins and identified a critical role of the C-terminus of the stathmin partner. Destabilizing this C-terminal region may represent an additional regulatory mechanism of the interaction with tubulin of stathmin proteins. More information: https://doi.org/10.1016/j.bbrc.2023.10.023 Contact: Benoît GIGANT <benoit.gigant@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 23, 2023 4:50 AM
|
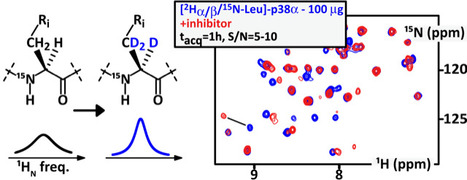
100 ug of kinase are now enough for its residue-scale NMR characterization
Kinases are exquisite drug targets, but usually too large for any NMR characterization at decent concentrations; we combined a number of innovative strategies enabling S/N~10 in 1h using less than 100 ug of protein. We aimed at designing and testing affordable strategies for improving NMR sensitivity on common-size drug targets, e.g. kinases of 40 kDa. This goes notably via novel isotope labeling schemes, which we sought to be as versatile as possible in terms of recombinant expression organism.
We evaluated the benefits of the deuteration on alpha- and beta-positions of amino acids for a typical middle size protein domain, namely the model 40 kDa-large kinase p38α.
We used cystathionine gamma-synthase (and new high-performance mutants thereof) to execute position-specific deuteration of free amino acids by H/D exchange. Then, we employed cell-free expression in bacterial extracts to avoid any scrambling and back-protonation of the tested isotopically labelled amino acids (Ala, Leu, Lys, Ser, Asp, Glu, Gly).
Altogether, these combined strategies yielded a ten-fold sensitivity increase, as compared to the former classical NMR approaches. It can be applied using recombinant expression in cell-free systems and in eukaryotic cells. This allows recording residue-resolved solution 1H-15N NMR spectra of 100 μg of purified p38α in one hour with S/N∼10, and a residue-specific characterization of drug binding.
We want to apply this approach for in-cell NMR studies in the future. More information: https://www.sciencedirect.com/science/article/pii/S2666441023000341?via%3Dihub Contact: Theillet Francois-Xavier <francois-xavier.theillet@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
September 20, 2023 8:48 AM
|
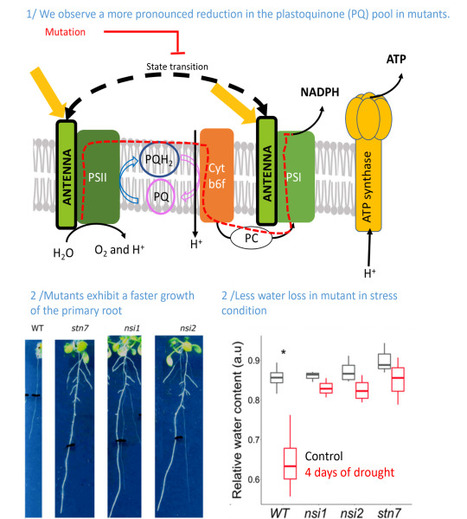
Identification of a new pathway potentially involved in plant resistance to drought
Light capture by the chlorophyll containing antenna and the reduction state of the photosynthetic electron transport chain affect root development. Identifying traits that exhibit improved drought resistance is highly important to cope with the challenges of predicted climate change. We investigated the response of state transition mutants to drought. Compared with the wild type, state transition mutants were less affected by drought. Photosynthetic parameters in leaves probed by chlorophyll fluorescence confirmed that mutants possess a more reduced plastoquinone (PQ) pool, as expected due to the absence of state transitions. Seedlings of the mutants showed an enhanced growth of the primary root and more lateral root formation. The photosystem II inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea, leading to an oxidised PQ pool, inhibited primary root growth in wild type and mutants, while the cytochrome b6 f complex inhibitor 2,5-dibromo-3-methyl-6-isopropylbenzoquinone, leading to a reduced PQ pool, stimulated root growth. A more reduced state of the PQ pool was associated with a slight but significant increase in singlet oxygen production. Singlet oxygen may trigger a, yet unknown, signalling cascade promoting root growth. We propose that photosynthetic mutants with a deregulated ratio of photosystem II to photosystem I activity can provide a novel path for improving crop drought resistance. More information: https://pubmed.ncbi.nlm.nih.gov/37614199/ Contact: Anja KRIEGER-LISZKAY <anja.krieger-liszkay@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
September 19, 2023 8:58 AM
|
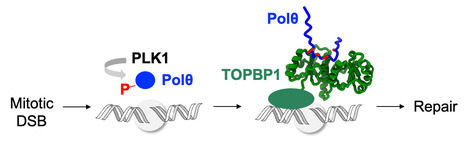
Phosphorylation of polymerase theta by PLK1 is essential for the repair of double-strand breaks in mitosis through a novel DNA repair pathway.
The DNA polymerase theta is phosphorylated by PLK1 in mitosis, binds to TOPBP1 and is recruited to double-strand breaks, in order to trigger repair through a novel DNA repair pathway, thus becoming a new target for the treatment of breast and ovarian cancers. DNA double-strand breaks (DSBs) are deleterious lesions that challenge genome integrity. In interphase, DSBs are mainly repaired by non-homologous end joining and homologous recombination. In mitosis, specific kinases inhibit these pathways through phosphorylation of essential DNA repair proteins. Here we show that one of these mitotic kinases, Polo-like kinase 1 (PLK1), activates the DNA polymerase theta (Polθ), which is then recruited through an interaction with TOPBP1 to mitotic DSBs. NMR analyses demonstrate that PLK1 phosphorylates a cluster of four serines in the central disordered region of Polθ, and that the phosphorylated motif directly interacts with the C-terminal domains of TOPBP1. The Artificial Intelligence based program AlphaFold consistently predicts that the Polθ region containing the cluster of four serines binds in a groove at the surface of the C-terminal domains of TOPBP1. Mutating these serines impairs recruitment of Polθ to mitotic DSBs and joining of the broken DNA ends. Polθ is essential for the repair of mitotic DSBs. Its role is even more crucial in cells that are deficient in homologous recombination, because these cells accumulate DSBs at the entry of mitosis, and loss of mitotic DSB repair by Polθ results in cell death. Our data explains why Polθ is synthetic lethal with homologous recombination deficiency, and reveals the critical importance of mitotic DSB repair in the maintenance of genome integrity. More information: https://www.nature.com/articles/s41586-023-06506-6 Contact: Sophie ZINN JUSTIN <sophie.zinn@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
July 19, 2023 10:17 AM
|
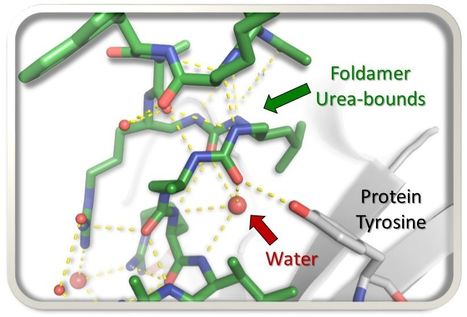
Unexpected binding modes of ureabased foldamers to a protein surface
Exploring uncharted territory, peptide-urea chimeras display surprising binding modes to the histone chaperone ASF1, rewriting the rules of interaction. In the search for foldamer inhibitors of the histone chaperone ASF1, the AMIG team of I2BC in collaboration with a team of Bordeaux University, explored the possibility of substituting four α-residues (≈ one helix turn) by 3-urea segments and scanned the sequence of a short α-helical peptide known to bind ASF1. By analysing the impact of the different foldamer replacements within the peptide chain, new binding modes of the peptide-urea chimeras to a protein target were discovered. More information: https://doi.org/10.1039/D3CC01891A Contact: Françoise Ochsenbein <francoise.ochsenbein@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
June 16, 2023 9:26 AM
|
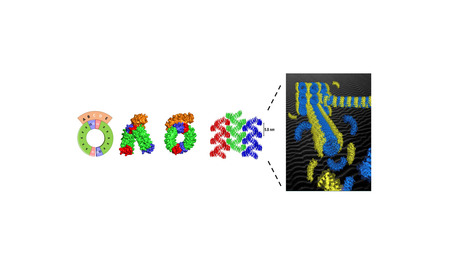
Self assembling structures from artificial proteins, or how to design protein origamis
Organized superstructures made from proteins such as microtubules, cilia or flagella are ubiquitous in living cells. How to design such highly organized superstructure from artificial protein components ? We describe a versatile strategy to create inducible protein assembly with predefined geometry. The assembly is mediated by a binding protein that staples identical protein bricks together in a predictable geometry. The first step was to select, among the alphaRep protein library, a protein able to interact with a specific area (the “back” surface) of another repeat protein used as a Bait. The structure of the “Bait”/”Back-binder” complex was then solved by the I2BC protein crystallography facility. Based on this structure, we designed a new protein, named the “Brick”, in which the surface elements of the Bait protein interacting with the “Back-binder” were split in two parts and appended at the two extremities of the “Brick” protein. This is made possible due to the extremely stable and modular organization of repeats proteins, such as alphaRep. In presence of the Brick protein, the Back-binder reconstitutes its cognate binding surface by joining the C end of one Brick protein to the N end of another Brick protein, in such a way that the surface elements of the two linked bricks adopt the same relative orientation as in the bait protein. Due to the twisted solenoid shape of the brick protein, the concatenation of many brick proteins leads to the formation of micrometer long super-helical filaments with a highly regular geometry. A very detailed structural characterization of the resulting assembly was conducted together with the Group of Erik Dujardin (CEMES Toulouse) using Negative stain electron microscopy (I2BC electron microscopy facility) , Small angle X ray scattering (F. Artzner, CNRS Rennes) , Cryoelectron Microscopy and Tomography (Integrative electron microscopy , Université de Toulouse). These efforts result in a detailed experimental description of the molecular organization and explain the higher order organization of the filaments by interdigitation of the back binders from neighboring filaments. The principles demonstrated here are potentially general and can be transferred to other types of affinity protein pairs derived either from natural or from computationally designed protein pairs and open routes toward the design and fabrication of multiscale protein origami with arbitrarily programmed shapes and chemical functions… More information: https://www.pnas.org/doi/10.1073/pnas.2218428120 Contact: Philippe Minard <philippe.minard@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
March 17, 2023 12:11 PM
|
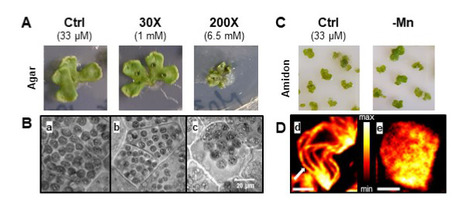
Manganese concentration affects chloroplast structure and the photosynthetic apparatus in Marchantia polymorpha.
Non-optimal manganese concentration affects photosynthetic electron transport and changes the ultrastructure of the chloroplast as revealed by super-resolution fluorescence microscopy. Manganese (Mn) is an essential metal for plant growth. It plays a major role in the chloroplast at the level of Photosystem II of the photosynthetic electron transport chain where it forms the Mn4CaO5 cluster, the site of water oxidation. Mn deficiency affects specifically photosynthesis while Mn excess is toxic for the plant. We studied Mn homeostasis in the liverwort Marchantia polymorpha whose simple morphology (absence of cuticle and archaic root system) allows a rapid and uncontrolled absorption. The determination of Mn in thalli and chloroplasts showed a high capacity of uptake (beyond even some metal hyperaccumulating plants) and resistance to stress, in particular thanks to some essential metabolites as shown by GC-MS analyses. In vivo super-resolution fluorescence microscopy (SRM) and transmission electron microscopy showed a change in chloroplast size and a disorganization of the thylakoid membranes. Mn deficiency increased the ratio of photosystem I compared to photosystem II. This alteration seems to favour cyclic electron flow around photosystem I and helps to protect the photosynthetic electron transport chain against photodamage. In conclusion, Marchantia polymorpha is an interesting model plant to study metal homeostasis and to be used by new analytical methods (MRS). In this organism, Mn plays an important role in thylakoid organization and photosynthetic electron transport. More: doi: 10.1093/plphys/kiad052 Contact: Anja Kieger-Liszkay <anja.liszkay@i2bc.paris-saclay.fr>
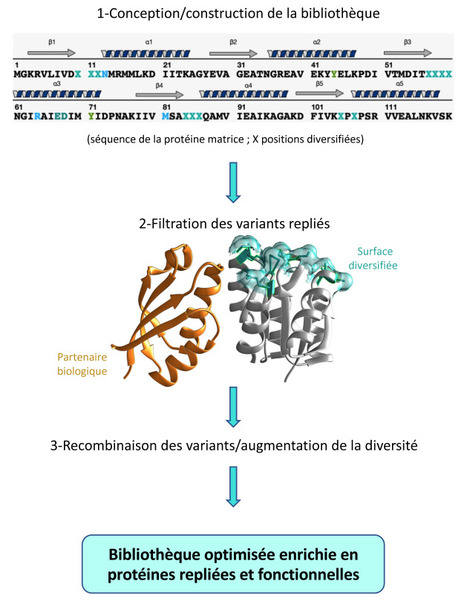
Les bibliothèques de protéines artificielles sont des outils remarquables pour identifier efficacement des protéines nouvelles interagissant spécifiquement avec n’importe quelle autre protéine cible choisie. Ces bibliothèques sont construites à partir d’une ossature protéique dont une partie de la surface est aléatoirement substituée. Plus cette surface est diversifiée, plus la bibliothèque peut être efficace. Cependant cette règle comporte une restriction majeure : la diversification tend généralement à augmenter la proportion de protéines de la bibliothèque devenues instables. Dans une étude publiée dans Scientific Reports, Marie Valerio-Lepiniec et ses collaborateurs de l’I2BC (CNRS/CEA/UPSaclay, Gif-sur-Yvette) ont donc développé une approche originale pour construire des bibliothèques qui soient à la fois très diverses et très stables. L’idée est de construire une bibliothèque diverse, puis de sélectionner au sein de celle-ci le sous-ensemble des protéines repliées et donc potentiellement stables et enfin de recombiner entre elles uniquement les séquences correspondantes. Les auteurs ont ainsi construit une bibliothèque en rendant hypervariable quatre boucles de la protéine utilisée comme ossature (CheY). Un partenaire protéique naturel de CheY interagissant avec une surface distincte de la surface diversifiée a été utilisé pour extraire de la bibliothèque initiale le sous-ensemble des variants repliés. Ensuite, une approche simple de recombinaison des séquences d'ADN codant ces variants « filtrés » a permis de restaurer une grande diversité. De cette façon, les chercheurs ont obtenu des protéines dont la stabilité moyenne est bien plus élevée que celle de la bibliothèque initiale. Cette bibliothèque améliorée a d’ores et déjà été utilisée avec succès pour sélectionner un inhibiteur spécifique de la Kaz-luciférase capable d'inhiber son activité de luminescence. Cet inhibiteur sera très utile pour concevoir de nouvelles familles de biosenseurs. Contact : marielle.valerio@i2bc.paris-saclay.fr
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
December 19, 2022 10:23 AM
|
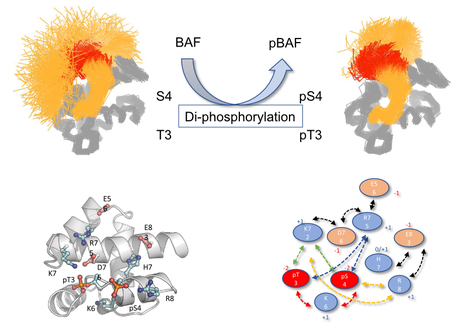
How Molecular Dynamics simulation and NMR show the effect of phosphorylation and pH on the conformational regulation of the intrinsically desordered N-terminal region of Barrier-to-Integration fact...
Mono- and, to a larger extent, di-phophorylation contribute to a gradual restriction of the conformationnal dynamics in the N-terminal region of the Protein BAF: A general mechanism for protein phosphorylation ? We studied the consequences of phosphorylation (the most frequent post-translational modification) on the conformation and dynamics of Barrier-to-Autointegration Factor (BAF) by numerical simulation and NMR. BAF is a highly conserved DNA binding protein, important for genome integrity. Its localization and function are regulated by phosphorylation. Previously published structures of BAF suggested that it is fully ordered, but our previous NMR analysis revealed that its N-terminal region is flexible in solution and that di-phosphorylation of residues S4 and T3 by the VRK1 kinase reduces this flexibility. Thanks to the computational means of TGCC via GENCI (Très Grand Centre de Calcul du CEA), we designed a molecular dynamics (MD) simulation protocol with multiple long duration (microsecond) simulations, each trajectory being characterized by the convergence of the conformational entropy of the simulated protein to ensure the completeness of the analyzed sets. This robust protocol allowed us to define the conformational ensembles accessible to the N-terminal region of BAF, whether unphosphorylated, mono-phosphorylated on S4, or di-phosphorylated on S4 and T3, and to reveal the interactions that help define these ensembles. We showed that the intrinsic flexibility of the N-terminal region of BAF is reduced by phosphorylation of the S4 residue and to a greater extent by di-phosphorylation of S4 and T3 residues. Thanks to the atomic description offered by MD simulation supported by by the NMR study of several BAF mutants, we detailed the mechanics of this restriction and described precisely the dynamic network of interactions responsible for the conformational restriction involving the phosphorylated residues pS4 and pT3. We also showed that the flexibility of the N-terminal region of BAF is influenced by ionic strength, pH and the essential role of the two negative charges carried by the phosphoryl groups for a substantial decrease in flexibility. Thus, the conformation of the intrinsically disordered N-terminal region of BAF appears to be highly adjustable, probably in connection with its various functions. This study also shows the contribution of numerical simulation in the context of structural biology for the study of the effect of post-translational modifications on protein structure and dynamics. More information: https://doi.org/10.1016/j.jmb.2022.167888 Contact: Philippe Cuniasse <philippe.cuniasse@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
November 24, 2022 8:48 AM
|
Artificials proteins, αReps, targeting the SARS-CoV-2 spike as antivirals
Specific binders targeting the SARS-CoV-2 spike protein were selected from the αRep library. Two combined αRep nanoligands neutralize SARS-CoV-2 variants and reduce infection severity in hamster. The coronavirus SARS-CoV-2 emerged as a human pathogen in China's Hubei province, causing severe respiratory diseases and pneumonia. Since the beginning of 2020, more than 600 million people have been infected with SARS-CoV-2 and more than 6.5 million have died during the COVID-19 crisis. Vaccines, although very effective in limiting severe forms, do not block the spread of the virus. Thus, reducing the multiplication of the virus in the nasal cavity is a promising approach to limit its spread.
In a new study published in the journal PLOS Pathogens, αRep artificial proteins were selected and used to prevent the virus from binding to its target cells and thus limit its multiplication. These proteins are able to recognize the SPIKE protein of the virus. More precisely, two αReps proteins, named F9 and C2 bind to the spike RBD domain with affinities in the nM range, and exhibit virus neutralization activity in vitro by recognizing two distinct sites. The combination of these αReps through covalent (F9-C2) or non-covalent (C2-foldon) linkages blocked the entry of the virus with a higher efficiency displaying affinity in the 0.1 nM range and EC50 of 8–18 nM for neutralization of SARS-CoV-2. These combination of αReps neutralize a broad spectrum of SARS-CoV-2 β, γ, δ and Omicron virus variants. Instillation of an αRep F9-C2 nanoligand in the nasal cavity of hamster effectively reduced virus replication and pathogenicity of SARS-CoV-2.
In addition to their strong antiviral capacity, these αReps can be very efficiently produced, at low cost, by recombinant protein expression technologies in bacteria. They are particularly robust, highly thermostable, and can be stored at room temperature. Taken together, the antiviral capacity associated with the attractive properties of these proteins are highly promising for the development of molecules able to reduce the pathology and spread of SARS-CoV-2 variants. More information: here Contact: Philippe Minard, <philippe.minard@i2bc.paris-saclay.fr>, Agathe Urvoas <agathe.urvoas@i2bc.paris-saclay.fr>, Marielle Valerio <marielle.valerio@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 24, 2022 4:45 AM
|
BSI@Paris-Saclay - nov 29, 2022
The Structural Integrative Biology (BSI) community is organising its first Scientific Day on Tuesday 29 November 2022 from 9:00 am to 5:00 pm at the I2BC (Auditorium of Bât21, CNRS campus, Gif-sur-Yvette) with the participation of the national infrastructures Soleil, Infranalytics and FRISBI and the European infrastructure Instruct-Eric. The objective of this day is to bring together the local BSI community of Paris-Saclay in order to foster new interactions. Speakers:
Harald SCHWALBE (Director of INSTRICT-ERIC)
Carine VAN HEIJENOORT (Director of Infranalytics)
Andy THOMPSON (Resp. scientist Synchrotron Soleil)
Stéphane BRESSANELLI (Resp. PF CryoEM, I2BC, FRISBI) And 5 oral presentations selected on abstract – Free but mandatory registration on the conference website
– Submission of abstracts (Posters and Oral Presentations) on the congress website.
The deadline for submission of abstracts for oral presentations is 21 October 2022. The selection will be made by the organising committee. We are looking forward to seeing many of you. Contacts:
julie.menetrey@i2bc.paris-saclay.fr and ewen.lescop@cnrs.fr Sponsors :
I2BC, ICSN, FRISBI, Infranalytics, Soleil, LM@W

|
Scooped by
I2BC Paris-Saclay
July 25, 2022 5:01 AM
|
Proteins are essential to the functioning of all organisms. Discover in this animated film what they are, what they are used for, how they are created, how they work, and how they are studied. All living things use proteins, make proteins, and are, in part, made of proteins. Proteins make up living things. They are in plant cells, in viruses and bacteria, in aquatic and terrestrial beings. They are in our muscles, in our blood, our hair, our tears, our saliva, our brain, our stomach. They also compose our food… These molecules are essential to the functioning of all organisms. What are they? What are they used for? How are they created? How do they work and how are they studied?
This animated film is intended for teachers and their students, and for all those who are curious about nature. Conceived in the form of 5 small chapters, its 2D and 3D animations will allow you to better understand what proteins are, what they are used for, how organisms make them according to their needs, how they function, how they interact with their environment, and finally what methods are used to study them.
|

|
Scooped by
I2BC Paris-Saclay
December 1, 2023 8:44 AM
|
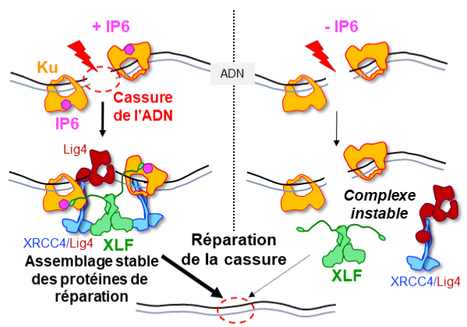
IP6 : an endogenous small molecule which promotes DNA repair.
Structural studies by X-ray cristallography and CryoEM combined with cellular analyses show the stabilizing role of IP6 in assembly of DNA double-strand break repair in human. IP6, inositol hexaphosphate or phytic acid, is a product of cellular metabolism that binds to numerous proteins whose functions it regulates. In an article published in the journal Nucleic Acids Research, scientists from I2BC (UMR 9198, CNRS/CEA/UPSaclay, Gif-sur-Yvette), from lPBS (CNRS/Université Paul Sabatier) and from Institut for Structural and Chemical Biology (University of Leicester, UK) reveal how IP6 stabilizes the assembly of a complex responsible for DNA break repair in humans. Many cancer therapies induce DNA double-strand breaks. A DNA double-strand break represents the most serious type of DNA damage to a cell, since it amounts to splitting a chromosome in two. The resulting chromosome fragments can be lost or lead to translocations if they are not quickly rejoined. Unrepaired breaks most often lead to cell death, a property exploited to eradicate tumor cells during radiotherapy or certain chemotherapies. Yet DNA double-strand break repair pathways exist, and their performance in tumors determines the efficacy of these therapies. The dominant system for repairing DNA double-strand breaks in human cells is Non-Homologous End Joining, or NHEJ, initiated by the Ku protein, which rapidly encircles the ends of the break and acts as a hub for the other proteins needed to weld these ends together. Using two techniques for studying proteins at atomic scale (crystallography and cryo-electron microscopy), the scientists discovered how a small molecule produced by the body and sometimes used as a dietary supplement, inositol-hexaphosphate (IP6) or phytic acid, binds to the Ku protein. The scientists then analyzed the effect of IP6 on break repair in human cells. They modified the Ku protein to block its binding to IP6, and thus understand how the latter acts. The Ku mutants revealed that the presence of IP6 on the Ku protein stimulates its binding to the XLF protein, an essential step in efficiently welding the DNA break (see the model below). IP6 thus plays a key role in stabilizing the large complex of NHEJ proteins required for DNA break repair. This fundamental work resolves the question raised two decades ago regarding the role of IP6 in DNA double-strand break repair. It also opens up therapeutic prospects by identifying a new region on the Ku protein that could be targeted by small molecules to block DNA break repair in tumor cells. More information: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkad863/7327077?login=false Contact: Jean-Baptiste CHARBONNIER <jb.charbonnier@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 23, 2023 4:58 AM
|
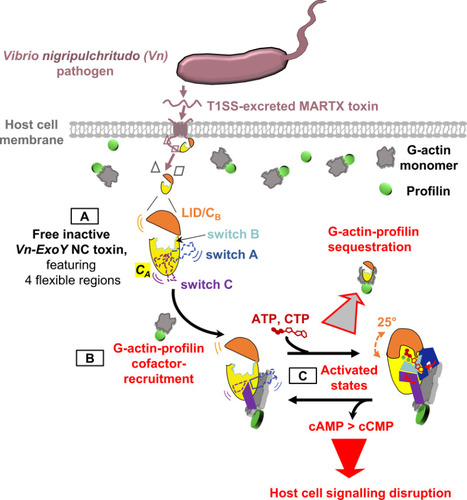
Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin
We reveal unprecedented mechanistic structural details of how the active site of bacterial Nucleotidyl Cyclase toxins is sequentially remodeled by cofactor and substrate binding and how they can accommodate both purine or pyrimidine nucleotides as substrate. Nucleotidyl cyclase (NC) enzymes are important ubiquitous enzymes that catalyze the production of cyclic nucleotides, crucial second messengers involved in numerous signaling pathways. ExoY-like toxins are bacterial virulence factors produced by Gram-negative γ- and β-proteobacteria. They belong to a broader family of bacterial NC toxins. When delivered into eukaryotic cells, NC toxins alter host cell signalling by overproducing both cyclic purine (cAMP, cGMP) and pyrimidine (cCMP, cUMP) nucleotides. To become potent NC enzymes inside host cells, they bind to specific host cofactors. The molecular and mechanistic details underlying the activation and catalytic specificities of NC toxins are only partially understood. ExoY NC toxins utilize either polymerized or monomeric (G-actin) actin as cofactor.
Here, we (a team at I2BC collaborating with scientists at ICSN, CNRS, UPS, Gif-Sur-Yvette, and at Institute Pasteur, Paris) investigated ExoY-like NCs that are selective for G-actin. Our in vitro investigations first revealed the physiological cofactor at work within host eukaryotic cells. These ExoYs activate by capturing and sequestering the cytoskeletal G-actin-profilin complex, disrupting its regulatory functions in the dynamic assembly of actin (Fig. 1, steps B-C).
Using X-ray crystallography at SOLEIL synchrotron, we have intercepted several structural snapshots along the ExoY activation pathway by G-actin-profilin. These structural data reveal unprecedented mechanistic details of how the active site of all NC toxins undergoes progressive remodeling upon cofactor and substrate binding (Fig. 1, step C). They reveal how NC toxins can accommodate purine or pyrimidine nucleotides as substrates and clarify crucial aspects of their catalytic purinyl and pyrimidinyl cyclase reaction. These structural insights into the multi-step activation of NC toxins may open new avenues to inhibit specifically this class of toxic bacterial NCs. More information: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1011654 Contact: Louis RENAULT <louis.renault@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 23, 2023 4:20 AM
|
Cyclope conference "Proteins are everywhere"
This conference is postponed to a later date. The next Cyclope conference organized by the CEA will be presented by Marie-Hélène Le DU at the INSTN at 8 p.m on the 24th of October 2023. The headline: “Proteins are everywhere". They are found in animals as well as plants, bacteria or viruses. They allow the regular beating of your heart, the digestion of your last meal, maintain your body at a constant temperature, participate in the fight against infections that threaten you and much more...
Proteins are everywhere life exists and science has studied them for decades, but they are so numerous and so diverse that their world has yet to be explored. We invite you to discover this exciting research in the company of Marie-Hélène Le Du, CEA researcher at the Institute of Integrative Cell Biology (DRF/Joliot/I2BC), with the participation of Rémi Ruedas, a doctoral student at I2BC.

|
Scooped by
I2BC Paris-Saclay
September 20, 2023 5:17 AM
|
Multiscale Transient Absorption Study of the Fluorescent Protein Dreiklang and Two Point Variants Provides Insight into Photoswitching and Nonproductive Reaction Pathways
Photoswitching and non-productive reaction pathways of the fluorescent protein 'Dreiklang' deciphered in a comprehensive time-resolved spectroscopic study. Dreiklang is a reversibly photoswitchable fluorescent protein used as a probe in advanced fluorescence imaging. It undergoes a unique and still poorly understood photoswitching mechanism based on the reversible addition of a water molecule to the chromophore. We report the first comprehensive study of the dynamics of this reaction by transient absorption spectroscopy from 100 fs to seconds in the original Dreiklang protein and its two point variants. The picture that emerges from our work is that of a competition between photoswitching and nonproductive reaction pathways. We found that photoswitching had a low quantum yield of 0.4%. It involves electron transfer from a tyrosine residue (Tyr203) to the chromophore and is completed in 33 ns. Nonproductive deactivation pathways comprise recombination of a charge transfer intermediate, excited-state proton transfer from the chromophore to a histidine residue (His145), and decay to the ground state via micro-/millisecond-lived intermediates. More information: https://pubs.acs.org/doi/10.1021/acs.jpclett.3c00431 Contact: Pavel MULLER <pavel.muller@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
July 19, 2023 10:22 AM
|
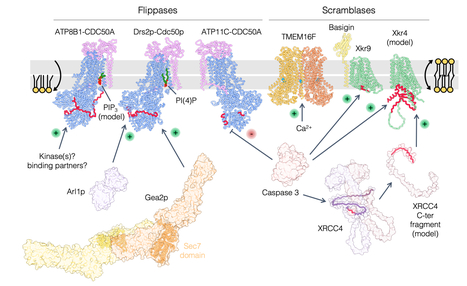
Phosphatidylserine transport in cell life and death
Lipids beyond membrane building blocks: recent advances in our understanding of phosphatidylserine distribution in eukaryotic cells and its role in infectious diseases. A prominent feature of cell membranes is the non-uniform distribution of the lipids they are made of. This is particularly true for the negatively charged glycerophospholipid phosphatidylserine (PS) which is enriched in the plasma membrane and in late secretory/endocytic compartments. In addition to the uneven distribution of PS between membranes, the transbilayer distribution (the enrichment of a given lipid in one membrane leaflet over the other) of PS is tightly regulated, as it is largely confined to the cytosolic leaflet of membranes, where it can mediate the recruitment of a diverse set of proteins. As such, PS controls a myriad of cell signaling pathways as well as membrane trafficking events. In some specific conditions, PS may however be ‘flipped’ toward the external leaflet of the plasma membrane, where it can act as an ‘eat-me’ signal for engulfment of apoptotic cells by macrophages or to promote synaptic pruning, which consists in the removal of synapses to establish proper connections during brain development. The exquisitely tailored transbilayer PS distribution is also crucial in host-pathogen interactions, as it is exploited by various intracellular pathogens to infect cells. In this review, we highlight recent findings on nonvesicular PS transport between membranes by lipid-transfer proteins at membrane contact sites, on PS flip-flop between membrane leaflets by lipid flippases and scramblases, and we discuss how perturbation of PS distribution can lead to disease and the role of PS in viral infection. More information: https://www.sciencedirect.com/science/article/pii/S0955067423000418?via%3Dihub Contact: Guillaume Lenoir <guillaume.lenoir@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
June 16, 2023 9:37 AM
|
How PAXX keeps DNA in check
We visualized the crucial steps of the molecular assembly allowing cells to repair breaks in their DNA, in particular those involving the PAXX protein. « It’s in my DNA » fine, but the information has to stay there without too much change, at the risk of losing our identity! Monitoring the health of DNA is the job of the many repair systems in our cells, each specialized in detecting and correcting a specific anomaly. A particularly serious threat is the breakage of the DNA double chain. For the cell, this is like a train whose tracks are suddenly interrupted: serious damage ahead! But the DNA is a fragile molecule that breaks easily, for example during flight under the effect of cosmic radiation, or upon exposure with radioactivity or X-rays, or simply because cells grow under reactive oxygen. The rail maintenance department in human cells is called the NHEJ, which coordinates many agents to detect breaks, protect them by a safety cord and finally weld them together. Of all the agents involved, the PAXX protein was the latest identified in 2015 but its role was not fully known. A B3S team in collaboration with a IPBS team in Toulouse (P Calsou) and a UK team in Bristol (A Chaplin) succeeded in reconstructing how PAXX functions in NHEJ. When the DNA is broken, repair begins with the ring-like Ku protein that very quickly encircles the ends of the break, serving as a mooring platform for the other proteins. Using two techniques that allow proteins to be visualized at the atomic scale (X-rays crystallography and cryo-electron microscopy), we captured a snapshot of the moment when PAXX docks with Ku and precisely identified their contact zones. They showed that PAXX works in tandem with XLF attached to the other side of Ku. When the binding sites are disrupted by mutations, the whole machinery is blocked, the break is no longer repaired, the train derails and the cell dies! In hyperactive cancer cells, the train travels at high-speed, explaining their special sensitivity to DNA breaks induced by radiotherapy ; molecules that would block the assembly of PAXX and/or XLF on DNA breaks could make it possible to increase the vulnerability of cancer cells and thus open up new therapeutic perspectives. The results of this research are therefore very promising and could lead to more effective therapies for cancer patients, although there is still work to be done to find and test these molecules. More information: https://pubmed.ncbi.nlm.nih.gov/37256950/ Contact: Jean-Baptiste Charbonnier <jb.charbonnier@i2bc.paris-saclay.fr> – Virginie Ropars <virginie.ropars@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
May 5, 2023 10:10 AM
|
4th meeting of the GdR MéDynA, October 3-6, 2023
Registration open until May 15, 2023: https://www.azur-colloque.fr/DR04/inscription/inscription/258 Self-assembled molecular systems are studied in various fields ranging from physics to biology, including chemistry, mathematics and materials science. While the properties (mechanical, biological) of complex molecular assemblies are already the subject of well-established scientific fields, the study of the dynamic phenomena leading to their formation and evolution over time remains a field in genesis. However, this is not only a field of fundamental scientific interest, but also of practical interest, such as for pathological assemblies or assemblies with therapeutic or (bio)technological purposes. For more details on MéDynA : https://medyna.cnrs.fr/ More information: here

|
Scooped by
I2BC Paris-Saclay
February 17, 2023 5:41 AM
|
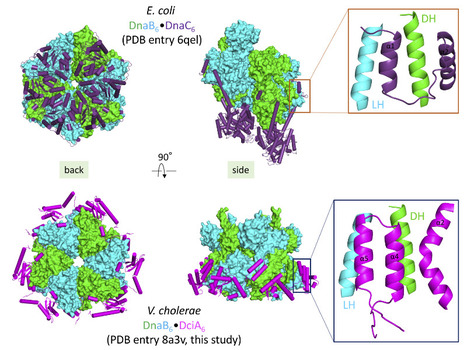
Convergent evolution of bacterial replicative helicase-helicase loader systems
The crystal structure of the DnaB•DciA complex from Vibrio cholerae reveals that the various helicase loaders share the same binding site on bacterial replicative helicases albeit with differences in their loading mechanism. Ring-shaped DnaB-family helicases play an essential role in bacterial genome replication, by unwinding DNA in front of replication forks. During the initiation step, their loading onto the replication origin depends on dedicated loader proteins. In particular, the well-characterized helicase loader DnaC from Escherichia coli cracks open the DnaB hexameric ring. However, DnaC, which is of phage origin, is an exception in the bacterial world. Instead, most bacteria contain the recently discovered and unrelated helicase loader DciA.
To understand how DciA assists the loading of DnaB onto DNA, the crystal structure of the complex from V. cholerae was determined. Strikingly, DciA binds to the periphery of the DnaB ring, in contrast to other known loaders such as DnaC, which oligomerize and are positioned at the back of the helicase. This discrepancy leads us to consider that the loading mechanism used by DciA, which has still to be elucidated, could differ from those previously described for other loaders. However, it is possible that a third partner, a protein or a nucleic acid, remains to be discovered to fully decipher the function of DciA.
Nevertheless, both DnaC and DciA target the same binding site: the conserved LH-DH module of DnaB helicases. This common binding site for various helicase loaders, as well as the fact that DciA and DnaC can be functionally interchanged in vitro, suggest convergent evolution of the different helicase-helicase loader systems. This could explain how phage loaders were able to efficiently hijack bacterial replicative helicases and replace the ancestral bacterial loader DciA several times during evolution. More information: https://doi.org/10.1107/S2059798323000281 Contacts: Hélène Wallbot, Sophie Quevillon-Cheruel, Stéphanie Marsin <helene.walbott@i2bc.paris-saclay.fr> <sophie.quevillon-cheruel@i2bc.paris-saclay.fr > <stephanie.marsin@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
January 16, 2023 9:32 AM
|
Accurate atomic model of the hepatitis E virus replication polyprotein
AlphaFold modelling of the multi-domain replication polyprotein of hepatitis E virus fosters insights on how this emerging pathogen generates new RNA viral genomes in infected cells. Structural biology, the field that produces atomic-level structures of biomacromolecules, has undergone several revolutions in the last ten years. The latest, and possibly the most far-reaching, is the development of artificial intelligence-based prediction of atomic structures of proteins from their sequence. The program AlphaFold has thus demonstrated an amazing capability to produce atomic models of proteins whose accuracy is actually on a par with that of experimental structures derived from X-ray crystallography or high resolution cryo-electron microscopy. Previously this could be achieved only for proteins with very high sequence identity to a protein of known structure. This development is thus particularly important for RNA viruses, whose sequences diverge fast and the proteins of which consequently never have high sequence similarity to other proteins of known structures.
Using AlphaFold, we modelled the hepatitis E virus (HEV) pORF1, the 1700-residue polyprotein that contains the necessary enzymatic activities to synthesise new HEV genomes in infected cells. We describe a five-domain protein with now clear boundaries for domains. The first of these, previously assigned to two distinct methyltransferase and Y domains, actually contains in a single domain both the activities necessary to cap the new viral RNAs (Met part), and interspersed membrane-interacting elements (Y part). We thus term this N-terminal domain MetY. The atomic structure of MetY is clearly homologous to the Chikungunya virus nsP1, despite very low sequence identity except in the methyltransferase active site. We propose a dodecameric assembly for MetY, as has been described experimentally for nsP1. MetY would thus form a pore allowing translocation from the membrane replication compartment to the cytosol coupled to capping of newly synthesised RNAs.
Most RNA viruses with genomes similar to HEV’s, such as hepatitis C virus (HCV), Chikungunya virus, Zika virus, or SARS-CoV-2, cleave their replication polyproteins into mature proteins with virally-encoded proteases. Our atomic model suggests that for HEV structural flexibility rather than proteolytic processing would serve to regulate pORF1 functions. The case of HCV has shown that direct-acting antivirals could cure chronic RNA virus infection, such as occurs in most HEV infections of immunocompromised patients. This work is a step forward in defining the molecular targets for such future HEV antivirals. More information: https://www.sciencedirect.com/science/article/pii/S0042682222002136 Contact: Sonia Fieulaine <sonia.fieulaine@i2bc.paris-saclay.fr> Thibault Tubiana <thibault.tubiana@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
November 24, 2022 8:59 AM
|
The 6th users meeting of the The French Infrastructure for Integrated Structural Biology (FRISBI) will be held on December 9th 2022 in Gif-sur-Yvette. Preliminary program and registration: https://frisbi.eu/network/user-community/ More information: here Contact: Sophie Zinn <sophie.zinn@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 24, 2022 5:07 AM
|
The BAF A12T mutation, associated to the Nestor–Guillermo progeria syndrome, disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures
The A12T mutation in BAF destabilizes the interaction with the nucleoskeletal lamin A/C and prevents robust repair of nuclear envelope ruptures in cells from patients with Nestor-Guillermo progeria. Nestor-Guillermo progeria syndrome (NGPS) is caused by a homozygous alanine-to-threonine mutation at position 12 (A12T) in barrier-to-autointegration factor (BAF). It is characterized by accelerated aging with severe skeletal abnormalities. BAF is an essential protein binding to DNA and nuclear envelope (NE) proteins, involved in NE rupture repair. The team of Dr Delphine Larrieu at the Cambridge Institute for Medical Research (UK), together with the team of Dr Sophie Zinn-Justin at I2BC and Dr Pierre Legrand at Synchrotron SOLEIL, assessed the impact of BAF A12T on NE integrity using NGPS-derived patient fibroblasts. The team of Dr Delphine Larrieu engineered the first NGPS isogenic cell lines by correcting the homozygous BAF A12T mutation in NGPS patient cells using CRISPR-Cas9 mediated genome editing. They observed a strong defect in lamin A/C accumulation to NE ruptures in NGPS cells, restored upon homozygous reversion of the pathogenic BAF A12T mutation. The team of Dr Sophie Zinn-Justin at I2BC, together with Dr Pierre Legrand at Synchrotron SOLEIL, demonstrated that, while the A12T mutation does not affect BAF 3D structure and phosphorylation by VRK1, it specifically decreases the interaction between BAF and lamin A/C. The team of Dr Delphine Larrieu revealed that the disrupted interaction does not prevent repair of NE ruptures but instead generates weak points in the NE that lead to a higher frequency of NE re-rupturing in NGPS cells. Altogether, we propose that this NE fragility could directly contribute to the premature aging phenotype in patients.

|
Scooped by
I2BC Paris-Saclay
September 21, 2022 12:14 PM
|
Third MéDynA plenary meeting (Assembly mechanisms and dynamics of self-organised protein-based complexes)
MéDynA third plenary meeting, 17-21 October, 2022 at Ile d'Oléron. More information here Contact: Stéphane Bressanelli <stephane.bressanelli@i2bc.paris-saclay.fr>
|



 Your new post is loading...
Your new post is loading...