Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
April 15, 8:44 AM
|
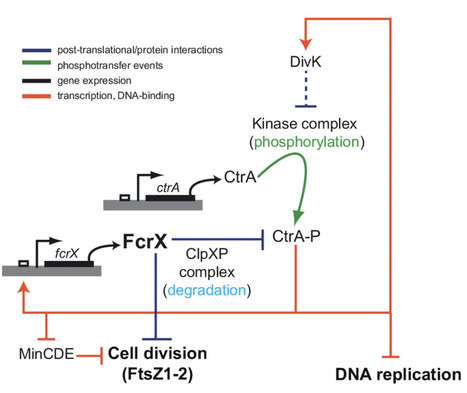
Sinorhizobium meliloti FcrX coordinates cell cycle and division during free-living growth and symbiosis by a ClpXP-dependent mechanism
In this study, a new essential player of cell cycle regulation has been characterized in the nitrogen fixing symbiont Sinorhizobium meliloti. During the nitrogen-fixing symbiosis between the alphaproteobacterium Sinorhizobium meliloti and the plant Medicago sativa (alfalfa), the interplay of molecular mechanisms governing cell cycle and bacteroid differentiation is a remarkable system that has still many details to be discovered. Here, we describe a bacterial cell cycle regulator, named FcrX, that controls two of the main essential players of cell cycle and bacteroid differentiation: the master regulator CtrA and the tubulin-like Z-ring component FtsZ. This essential factor is potentially participating with the degradosome complex driving the proteolysis of those two important regulators. Constitutive expression of FcrX during nodule development shows an increase of plant biomass, opening interesting paths in the amelioration of biological nitrogen fixation for a sustainable agriculture. More information : https://doi.org/10.1073/pnas.2412367122 Contact : Emanuele Biondi - emanuele.biondi@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
February 17, 5:00 AM
|
ChemPhysBio2025 : Registrations Now Open!
We’re excited to announce that registrations are now open for the Second Interdisciplinary Summer School: ChemPhysBio2025 – Imaging Biology: Interdisciplinary Tools and Techniques to Unravel Cellular Dynamics – taking place June 16–20, 2025, at Université Paris-Saclay! This unique event bridges chemistry, physics, and biology, offering participants the opportunity to explore advanced photonic imaging techniques for unraveling cellular dynamics. We’re also thrilled to welcome two distinguished guest lecturers: - Julie Karpenko (Laboratory for Therapeutic Innovation, University of Strasbourg)
- Balázs Enyedi (Department of Physiology, Semmelweis University, Hungary)
What to expect? - Inspiring lectures and research seminars
- Interactive interdisciplinary workshops
- Hands-on experience in cutting-edge research labs
Who should apply?
Master’s students, PhD candidates, postdocs, and junior researchers who are newcomers to the interdisciplinary field and eager to deepen their knowledge, expand their skill sets, and build meaningful collaborations. With only 20 spots available, don’t miss your chance to join us!
Apply now: https://chemphysbio2025.sciencesconf.org/
Portrait Jeune Chercheur – Christopher Dane Fage, Chercheur en biochimie structurale
Christopher Dane Fage is a structural biochemist who, in January of 2024, joined the Institute for Integrative Biology of the Cell - I2BC (CNRS/CEA/UPSaclay, Gif-sur-Yvette) as a CNRS CRCN to lead the team Structures and functions of hybrid natural product assembly lines. His research seeks to understand and exploit the biosynthesis of medically relevant bacterial metabolites by multi-family enzyme systems. In 2014, Chris earned his PhD from the University of Texas at Austin (USA) in the team of Adrian Keatinge-Clay, where his interest in the structural biochemistry of polyketide synthases (PKSs) crystallized (these multi-domain, multi-module enzymatic assembly lines build complex molecules from simple building blocks through the catalytic chemistry of fatty acid synthases (FASs)). For example, from the spinosyn insecticide pathway, he investigated a Diels-Alderase-like enzyme (catalyzes a reaction widely used in chemical synthesis) and a “dimerization element” (organizes the modular architecture of many PKSs). Subsequently, as a postdoc at Philipps-Universität Marburg (Germany) with Mohamed Marahiel, Chris focused on other types of natural product-forming enzymes, such as non-ribosomal peptide synthetases (NRPSs; these assembly line systems incorporate different building blocks than PKSs using different catalytic domains). For example, he initiated an integrative structure-function study on the inter-subunit “COM” interfaces needed for efficient peptide formation in many NRPSs. He also studied the properties, detection, biosynthesis, and tailoring of the ribosomally synthesized and post-translationally modified lasso peptides, which adopt a remarkably stable lariat knot-like fold. From 2017, Chris joined the teams of Greg Challis and Józef Lewandowski at the University of Warwick (UK). There, he investigated trans-AT PKSs, which employ a wide variety of standalone enzymes in place of their subunit-embedded counterparts, as well as hybrid PKS-NRPS systems, which merge the catalytic chemistry of PKSs and NRPSs – thanks to their peculiarities, these machineries build exceptionally diverse compounds. He also clarified the mode of action of the kalimantacin antibiotics against the human pathogen Staphylococcus aureus. In a brief departure from natural products, Chris worked at the Centre for Medicines Discovery at the University of Oxford (UK) during 2022-2023, where he characterized the structures of three enzymes associated with type 2 diabetes mellitus. He also developed a high-throughput assay for screens against libraries of tens of thousands of inhibitors. Since arriving at the I2BC, Chris has been pursuing structure-guided studies towards the rational reprogramming of hybrid FAS-NRPS-PKS systems, with particular interest in those of the kalimantacin (antibacterial) and bleomycin (anticancer) drugs. “The true delight is in the finding out rather than in the knowing.” - Isaac Asimov -> Contact : christopher.fage@i2bc.paris-saclay.fr
Via Life Sciences UPSaclay
Le prochain café se tiendra le vendredi 22 novembre 2024 en visioconférence de 13h30 à 14h avec la participation de Denis Faure (I2BC) qui nous parlera sur « The versatile lifestyle of the plant pathogen Agrobacterium tumefaciens » Pour assister en visioconférence : ICI
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
October 17, 2024 6:00 AM
|
Symposium Archaea October 17-18 2024

|
Scooped by
I2BC Paris-Saclay
October 1, 2024 6:11 AM
|
Symposium Archaea 2024, 17-18 october at I2BC
Want to learn more about the third domain of life, the Archaea? Join us at I2BC for an annual symposium gathering French research community studying archaea. The Archaea 2024 symposium, organised by the SFBBM's GT-Archées, is intended to bring together the entire French scientific community to discuss and exchange ideas on unifying themes relating to the importance and place of Archaea, the third domain of living organisms, in ecosystems, evolution and the fundamental processes of life. The scientific community is seeking to elucidate the fundamental molecular mechanisms at work in these microorganisms, the diversity of their viruses, and the adaptive limits of living organisms in relation to the environment. Archaea have been the subject of study from a variety of disciplinary perspectives, including those of biochemists, microbiologists, evolutionists, molecular biologists and also from the perspective of industry. Archaea are now being regarded as essential models for understanding the processes that underpin the functioning of living organisms. more information: https://archaea2024.sciencesconf.org/ Contact: tamara.basta@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 23, 2024 10:24 AM
|
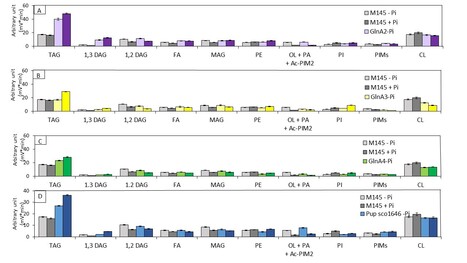
Impact of the Deletion of Genes of the Nitrogen Metabolism on Triacylglycerol, Cardiolipin and Actinorhodin Biosynthesis in Streptomyces coelicolor.
Analysis of the consequences of the deletion of genes involved in nitrogen metabolism on the content of phospholipids or of storage lipids of Streptomyces coelicolor illustrates the complex links exiting between carbon, nitrogen and phosphate metabolisms. Since nitrogen (N) limitation is known to be an important trigger of storage lipid / triacylglycerol (TAG) accumulation, in most microorganisms, we assessed the global lipid content of 21 strains derived from Streptomyces coelicolor deleted for genes involved in N metabolism. These strains were grown in the classical R2YE medium that is N and phosphate (P) limited. Seven of these strains had a higher TAG content than the original strain. These strains were either deleted for genes encoding proteins involved 1) in polyamine degradation (GlnA2/SCO2241, GlnA3/SCO6962, GlnA4/SCO1613); 2) in protein degradation (Pup/SCO1646); 3) in the regulation of nitrogen metabolism (GlnE/SCO2234 and GlnK/SCO5584) or 4) encoding the global regulator DasR/SCO5231 that controls negatively the degradation of N-acetylglucosamine, a constituent of peptidoglycan. Five of these strains, with the exception of the glnA2 and pup mutants, had a lower cardiolipin (CL) content than the original strain. This suggested that in these strains the biosynthesis of TAG, molecule devoided of (P) groups is prevalent over that of CL that bears 2 (P) groups, thus allowing higher (P) availability. The deletion of pup, glnA2, glnA3, and glnA4 was correlated, in Pi limitation, with an increase in the total production of the blue polyketide actinorhodin (ACT), whereas ACT production was strongly reduced in the glnA3 mutant in (P) proficiency and was totally abolished in the dasR mutant in both (P) conditions. Altogether, our data suggest that N limitation linked to the deletion of genes mentioned above results into high oxidative stress that triggers storage of acetylCoA as TAG to reduce the activity of the oxidative metabolism, generator of oxidative stress, as well as the biosynthesis of ACT that has anti-oxidant properties. At last, the reduced ACT biosynthesis of the glnA3 mutant, is thought to be due to its high spermine and spermidine content, molecules known to protect the cell against oxidative stress. More information: doi.org/10.3390/microorganisms12081560 Contact: Marie-Joëlle VIROLLE marie-joelle.virolle@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 26, 2024 8:09 AM
|
From Molecules to Organisms: Advancing Labelling, Imaging and Analysis of Biological Samples

|
Scooped by
I2BC Paris-Saclay
March 25, 2024 9:58 AM
|
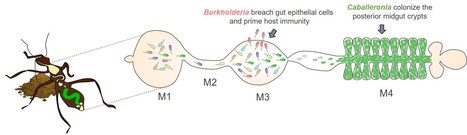
Members of gut microbiota prime insect immunity
Burkholderia bacteria from the gut microbiota in the bean bug insect can cross the epithelium of the gut and trigger a protective systemic immune response that has no adverse effects on the insect fitness but confers a general protection against pathogens. Insects lack acquired immunity and were thought to have no immune memory, but recent studies reported a phenomenon called immune priming, wherein sub-lethal dose of pathogens or non-pathogenic microbes stimulate immunity and prevent subsequential pathogen infection. Although the evidence for insect immune priming is accumulating, the underlying mechanisms are still unclear. The bean bug Riptortus pedestris acquires its gut microbiota from ambient soil and spatially structures them into a multispecies and variable community in the anterior midgut and a specific, mono-species Caballeronia symbiont population in the posterior region. We demonstrate that some particular Burkholderia strains colonizing the anterior midgut, stimulate systemic immunity by penetrating gut epithelia and migrating into the hemolymph. This hemolymph colonization and activated immunity, consisting of a humoral and a cellular response, has no negative effect on the host fitness, but on the contrary protected the insect from subsequent infection by pathogenic bacteria. Interruption of contact between the Burkholderia and epithelia of the gut weakened the host immunity back to pre-infection levels and made the insects again vulnerable to microbial infection, demonstrating that persistent acquisition of environmental bacteria is important to maintain an efficient immunity. This suggests that priming is only activated when it might be most helpful, e.g. when pathogen encounters are most likely in a microbially rich environment. Together, these findings not only highlight a role of environmental microbes in shaping insect immunity but also put a new, symbiotic perspective on bacterial intestinal barrier breaching and hemolymph colonization, which has been generally viewed only as a pathogenic phenomenon. More information: https://doi.org/10.1073/pnas.2315540121 Contact : Peter MERGAERT <peter.mergaert@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
March 18, 2024 11:38 AM
|
"Jeudis de la Recherche" at I2BC
As part of the "Jeudis de la Recherche" organized by the COMPAS (Paris-Saclay University), the CNRS and the town of Gif sur Yvette, Sylvie Lautru's team (I2BC, Microbiology Department) welcomed around twenty people to take part in 5 workshops on the theme of "Antibiotics and the microbial response: a never-ending battle". Thanks to the very interactive format, participants who were very interested in this topical issue, asked many questions and exchanged a lot with the research team. https://www.ville-gif.fr/215/que-faire-a-gif/culture/conferences/jeudis-de-la-recherche.htm

|
Scooped by
I2BC Paris-Saclay
February 19, 2024 10:19 AM
|
Prize "Relève de l'étoile" for the excellence of the research work carried out by Alexia Royer
Alexia Royer received the prize "Relève de l'étoile" for her article “Clostridioides difficile S-Layer Protein A (SlpA) Serves as a General Phage Receptor” published in Microbiology Spectrum in February 2023. Antibiotics have been used for more than 80 years to treat bacterial infections, but bacteria have developed resistance to these treatments, making them difficult to eradicate. One of the side effects of antibiotics is that they tend to kill a broad spectrum of bacteria, including “good bacteria” important for the proper functioning of the human body. The resulting bacterial imbalance, called dysbiosis, will open the door to opportunistic bacteria such as Clostridioides difficile. This bacterium is the main cause of severe diarrhea in industrialized countries. As the infection is now treated with antibiotics, which are themselves responsible for the infection, many relapses are reported. In the laboratories of Professor Louis-Charles Fortier and Professor Olga Soutourina, we are interested in phage therapy, which is a treatment based on the use of bacteria-killing viruses called bacteriophages. Phages have the advantage of being specific and targeting a species or a few bacterial strains unlike antibiotics. Thus, they could be used as an alternative or complementary therapy to antibiotics to treat patients in therapeutic failure. The key step to set up a phage therapy is to understand the mechanism of recognition of the bacteria by phages. For a phage to attack and kill a bacterium, it must first attach to a receptor on its surface. In the article, we identified the SlpA protein as the receptor on the surface of the bacteria that is targeted by phages. We have shown that the infection specificity of phages is linked to the presence of different forms of SlpA, called isoforms, each strain expressing one isoform. We also identified a domain of SlpA, named D2, playing a role in receptor recognition by certain phages. These data are essential because they improve our understanding of phage/bacteria interactions and make phage therapy possible one day against C. difficile infections. https://frq.gouv.qc.ca/histoire-et-rapport/releve-etoile-jacques-genest-decembre-2023/ Alexia ROYER <alexia.royer@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
February 19, 2024 10:10 AM
|
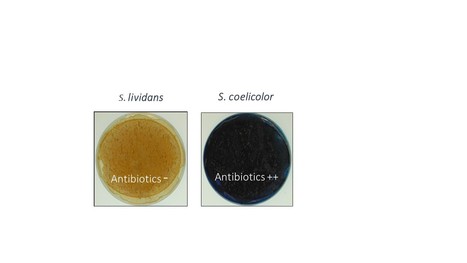
The stringent response is strongly activated in the high antibiotic producer, Streptomyces coelicolor
The stringent response controls positively antibiotic biosynthesis. Antibiotics are thus part of the stringent response. In most bacteria, the stringent response (SR) was initially characterized as a response to nitrogen (N) limitation resulting into the depletion of aminoacylated tRNAs leading to the stalling of ribosomes on mRNA. The recruitment of the ppGpp synthetase, RelA, at the stalled ribosomes activates (p)ppGpp synthesis from GTP. ppGpp that is the mediator of the SR controls negatively, at the transcriptional and translational levels, the expression of most ribosomal proteins leading to the down regulation of the translational process and thus of growth.
The model strains Streptomyces coelicolor (SC) and Streptomyces lividans (SL), strong and weak producers of the same antibiotics, respectively, were grown in condition of phosphate (Pi) limitation or proficiency and the abundance of proteins of their translational apparatus was compared. This study revealed that the expression of RelA was induced in Pi limitation suggesting that, besides N limitation, Pi limitation also contributes to the triggering of the SR. Interestingly, most proteins of the translational apparatus had a similar or slightly higher abundance in SL than in SC, in Pi limitation whereas most of these proteins were far more abundant in SL than in SC, in Pi proficiency. This indicated an alleviation of the SR in Pi proficiency in SL, but not in SC. This suggested an alteration of Pi up-take and/or Pi-mediated regulation in SC whose molecular basis remain to be elucidated.
Interestingly, the production of specialized metabolites in SC (CDA, RED and ACT) is usually concomitant of phases of growth slow down and it is known that ppGpp controls positively the expression of their biosynthetic pathways. Their production could thus be considered as part of the SR. Indeed, these metabolites were proposed to regulate negatively, through different processes, the energetic metabolism and thus the generation of ATP, in SC, a process that might contribute to the slower growth rate of SC compared to SL. More information: https://www.sciencedirect.com/science/article/pii/S0923250823001547?via%3Dihub Contact: Marie-Joëlle VIROLLE <marie-joelle.virolle@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 1, 2023 8:47 AM
|
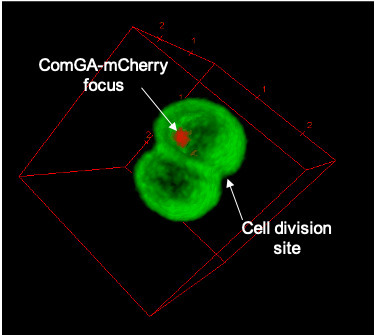
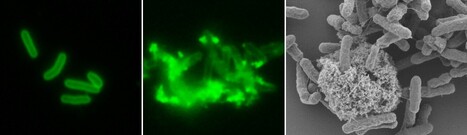
Natural transformation and cell division delay in competent Staphylococcus aureus
We hypothesize that S. aureus competent cells would initiate and then block cell division to ensure the success of natural transformation before the final constriction of the cytokinetic ring.. Genetic competence for natural transformation, considered one of the three main mechanisms leading to horizontal gene transfer in bacteria, is able to promote evolution, through genomic plasticity, and foster antibiotic resistance and virulence factors spreading. Conserved machinery and actors required to perform natural transformation have been shown to accumulate at different cellular localizations depending on the model organism considered. In this study, we investigate the transformation apparatus composition, localization, and dynamics in the human pathogen Staphylococcus aureus. We particularly show that most of the natural transformation actors co-localize in clusters. We also reveal that the localization of natural transformation proteins is dynamic, following the cell cycle. Ultimately, the natural transformation apparatus is preferentially established in the vicinity of the division septum. All these results demonstrate that DNA binding, uptake, and recombination are spatially and temporally coordinated to ensure S. aureus natural transformation. Finally, we hypothesize that S. aureus competent cells would initiate and then block cell division to ensure the success of natural transformation before the final constriction of the cytokinetic ring. More information: https://journals.asm.org/doi/10.1128/spectrum.02807-23 Contact: Nicolas MIROUZE <nicolas.mirouze@i2bc.paris-saclay.fr>
|

|
Scooped by
I2BC Paris-Saclay
April 15, 8:38 AM
|
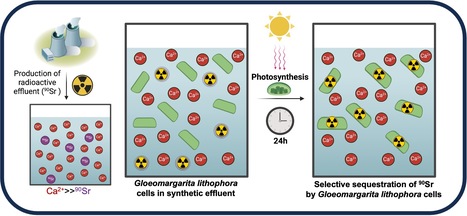
The cyanobacterium G. lithophora is a promising organism for effective bioremediation of waters contaminated with 90Sr
The unicellular cyanobacterium Gloeomargarita lithophora is able to stably sequester and withstand 90Sr and efficiently remove this hazardous anthropogenic radionuclide from aqueous solutions, including a synthetic nuclear effluent. Strontium (Sr) is an alkaline earth metal commonly occurring in nature. In aqueous environments, it prevails as Sr2+ ions form chemically similar to Ca2+. In most organisms, the non-selective Sr intake primarily comes from water and food. In human, it is mainly localized in bones and dental enamel and is involved in the control of bone formation. Radioactive isotopes such as 90Sr are artificially produced by nuclear fission. 90Sr, a b-emitter with a half-life of 28.8 years, is one of the most hazardous anthropogenic radionuclides. It has been massively released into the environment during nuclear weapons testing and nuclear reactor accidents, such as those at Chernobyl and Fukushima. It is also found in radioactive water effluents produced by nuclear power plants, requiring specific treatment before being discharged. In humans, its accumulation in bone tissues increases the risk of cancers. Current physico-chemical techniques for remediating 90Sr traces from effluents are costly and can exhibit a low selectivity for Sr over Ca. Therefore, there is an incentive to develop an alternative method of remediation. In this study, we demonstrate that the cyanobacterium Gloeomargarita lithophora can remove more than 90% of the 90Sr activity that could be found in a nuclear plant effluent within 24 hours. This process occurs through two steps: a first rapid and passive phase of 90Sr sorbtion to the cell surface, followed by an active phase of 90Sr accumulation within the cells, partially driven by photosynthesis. We also showed that this cyanobacterium is able to stably sequester and withstand 90Sr and to efficiently remove this hazardous radionuclide from aqueous solutions, including a synthetic nuclear effluent. These results highlight Gloeomargarita lithophora as a promising solution for effective bioremediation of water contaminated with 90Sr thereby safeguarding the well-being of our ecosystems.
More information : https://doi.org/10.1016/j.jhazmat.2025.138155 Contact : Corinne CASSIER-CHAUVAT Corinne.CASSIER-CHAUVAT@cea.fr

|
Scooped by
I2BC Paris-Saclay
February 11, 8:55 AM
|
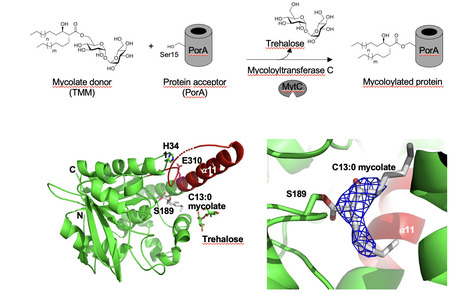
Synthetic mycolates derivatives to decipher protein mycoloylation, a unique post-translational modification in bacteria
In vitro reconstitution of bacterial protein mycoloylation. Protein mycoloylation is a newly characterized post-translational modification (PTM) specifically found in Corynebacteriales, an order of bacteria that includes numerous human pathogens. Their envelope is composed of a unique outer membrane, the so-called mycomembrane made of very-long chain fatty acids, named mycolic acids. Recently, some mycomembrane proteins including PorA have been unambiguously shown to be covalently modified with mycolic acids in the model organism Corynebacterium glutamicum by a mechanism that relies on the mycoloyltransferase MytC. This PTM represents the first example of protein O-acylation in prokaryotes and the first example of protein modification by mycolic acid. Through the design and synthesis of trehalose monomycolate (TMM) analogs, we prove that i) MytC is the mycoloyltransferase directly involved in this PTM, ii) TMM, but not trehalose dimycolate (TDM), is a suitable mycolate donor for PorA mycoloylation, iii) MytC is able to discriminate between an acyl and a mycoloyl chain in vitro unlike other trehalose mycoloyltransferases. We also solved the structure of MytC acyl-enzyme obtained with a soluble short TMM analogs which constitutes the first mycoloyltransferase structure with a covalently linked to an authentic mycolic acid moiety. These data highlight the great conformational flexibility of the active site of MytC during the reaction cycle and pave the way for a better understanding of the catalytic mechanism of all members of the mycoloyltransferase family including the essential Antigen85 enzymes in Mycobacteria. more information: https://doi.org/10.1016/j.jbc.2025.108243 Contact: Florence CONSTANTINESCO-BECKER <florence.constantinesco@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
November 26, 2024 3:25 AM
|
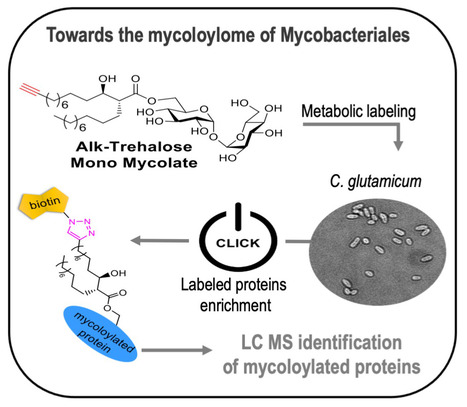
Bioorthogonal monomycolate of trehalose disclosed the O-mycoloylation of mycoloyltransferases and other cell envelope proteins in C. glutamicum
Discovering the mycoloylome of Corynebacterium glutamicum : a new type of lipidated-modified proteins. Protein mycoloylation is a lipidation pathway exclusively observed in Mycobacteriales, an order of bacteria that includes several human pathogens. These bacteria possess a distinctive outer membrane, known as the mycomembrane, composed of very long-chain fatty acids called mycolic acids. It has been demonstrated that a few mycomembrane proteins undergo covalent modification with mycolic acids in the model organism Corynebacterium glutamicum, through the action of mycoloyltransferase MytC. This PTM represents the first example of protein O-acylation in prokaryotes and also the first example of protein modification by mycolic acid. Here, we have developed a unique bioorthogonal mycolate donor featuring the natural mycolic acid pattern, enabling direct unambiguous transfer of the lipid moiety to its acceptors and efficient metabolic labeling and enrichment of MytC protein substrates. Mass spectrometry analysis of the labeled proteins and comparative proteomic analysis of the cell envelope proteome between wild-type and ∆mytC strains identified an unbiased list of 21 proteins likely mycoloylated in the cell. The robustness of our approach is demonstrated by the successful biological validation of mycoloylation in 6 random candidate proteins within wild-type cells, revealing the characteristic profile of proteins modified with natural mycolates. These findings provide interesting insights into the significance of this new lipidation pathway and pave the way for understanding their function, especially concerning the mycoloyltransferase family that includes the essential Antigen85 enzymes in Mycobacteria. More information: https://doi.org/10.1021/acschembio.4c00502. Contact: Cécile LABARRE <cecile.labarre@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
October 17, 2024 6:06 AM
|
Plant Science Day October 17th

|
Scooped by
I2BC Paris-Saclay
October 1, 2024 6:15 AM
|
The I2BC will be present at the Fête de la Science on 5 and 6 October 2024 at ENS Paris-Saclay (4 avenue des sciences- Gif sur Yvette) on the theme ‘Ocean of knowledge’. The Microbiology Department and the Crystallography Platform will each have a stand entitled ‘Can bacteria swim?’ and ‘Making and observing protein crystals’, where the public can carry out experiments, look under a microscope and take part in games. more information: https://www.fetedelascience.fr/ Contact: catherine.grandclement@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 1, 2024 6:05 AM
|
Awarding of the 2024 Haüy-Lacroix Thesis Prize to Chloé Truong
The SFMC (Société Française de Minéralogie et de Cristallographie) jury has decided to award the Haüy-Lacroix 2024 prize to Chloé TRUONG for her thesis work, "In search of biosignatures of hyperthermophilic archaea". Chloé TRUONG’s thesis, entitled “In search of biosignatures of hyperthermophilic archaea”, was carried out at the Institut de Minéralogie, de Physique des Matériaux et de Cosmochimie (Museum National d’Histoire Naturelle de Paris) under the supervision of François Guyot, Aurore Gorlas and Sylvain Bernard. The aim of her thesis was to determine whether the black smoker vents at ocean ridges, which expel warm, metal-rich water, can host hyperthermophilic life, i.e. organisms that develop in water at over 100°C. To do this, Chloé TRUONG adopted a multidisciplinary approach combining experimental mineralogical and microbiological studies in the laboratory and the characterisation of natural samples. This thesis provides major results on the formation and evolution of mineral phases such as pyrite, demonstrating that its presence can result from the activity of microorganisms. The identification of such mineral biosignatures will enable a systematic search for traces of life in modern and fossil black smokers. more information: https://sfmc-fr.org/?p=3516&lang=fr Contact: aurore.gorlas@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 26, 2024 8:21 AM
|
SYNTERUPTOR : mining genomic islands for non-classical specialized metabolite gene clusters
In search of the hidden gene cluster: Synteruptor, a new tool for identifying bacterial genomic islands and exploring their content in the quest for new specialized metabolism gene clusters. Microbial specialized metabolite biosynthetic gene clusters (SMBGCs) are a formidable source of natural products of pharmaceutical interest. With the multiplication of genomic data available, very efficient bioinformatic tools for automatic SMBGC detection have been developed. Nevertheless, most of these tools identify SMBGCs based on sequence similarity with enzymes typically involved in specialised metabolism and thus may miss SMBGCs coding for undercharacterised enzymes. In this article, we present Synteruptor (https://bioi2.i2bc.paris-saclay.fr/synteruptor), a program that identifies genomic islands, known to be enriched in SMBGCs, in the genomes of closely related species. With this tool, we identified a SMBGC in the genome of Streptomyces ambofaciens ATCC23877, undetected by antiSMASH, the well-known and most used tool for SMBGC identification, prior to antiSMASH 5. We experimentally demonstrated that this SMBGC directs the biosynthesis of two metabolites, one of which was identified as sphydrofuran. Synteruptor is also a valuable resource for the delineation of individual SMBGCs within antiSMASH regions that may encompass multiple clusters, and for refining the boundaries of these SMBGCs. More information: https://doi.org/10.1093/nargab/lqae069 Contact: Sylvie LAUTRU <sylvie.lautru@i2bc.paris-saclay.fr> & Olivier LESPINET <olivier.lespinet@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
July 2, 2024 10:14 AM
|
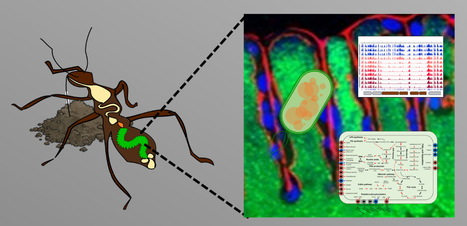
Hundreds of antimicrobial peptides create a selective barrier for insect gut symbionts
Antimicrobial peptides shape the gut microbiota biogeography in insects. The microbioata is usually not homogeneously dispersed in the animal gut but spatially structured in microenvironments. The microbiota in the gut of the bean bug Riptortus pedestris displays a sharp divide between the anterior and posterior midgut with a multispecies bacterial community in the anterior region and a specific, mono-species Caballeronia symbiont population in the posterior region. In this collaborative work between I2BC teams, the Next-generation sequencing plateform, the scanning electron microscopy plateform of MICALIS and a team from the AIST, Sapporo Japan, we found that this insect deploys in the midgut an arsenal of several hundreds of antimicrobial peptides. These peptides have antimicrobial activity against diverse bacteria but posterior midgut symbionts have elevated resistance while mutants of these symbionts in resistance genes have reduced capacity to colonize the posterior midgut. The peptides create thus a selective environment restricting the type of bacteria from the anterior midgut microbiota that have a chance to establish in the posterior midgut. This finding highlights a mechanism that contributes in the construction of an exclusive niche for beneficial gut symbionts. More information: https://www.pnas.org/doi/10.1073/pnas.2401802121 Contact: Peter Mergaert peter.mergaert@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
March 25, 2024 6:16 AM
|
Cooperation between two modes for DNA replication initiation in the archaeon Thermococcus barophilus
Demonstration that diverse physiological states influence the mode of DNA replication initiation in the archaeon Thermococcus. The mechanisms underpinning the replication of genomic DNA have recently been challenged in Archaea. Indeed, the lack of origin of replication has no deleterious effect on growth, suggesting that replication initiation relies on homologous recombination. Recombination-dependent replication (RDR) appears to be based on the recombinase RadA, which is of absolute requirement when no initiation origins are detected. The origin of this flexibility in the initiation of replication and the extent to which it is used in nature are yet to be understood. We combined deep sequencing and genetics to elucidate the dynamics of oriC utilization according to growth phases. We discovered that in Thermococcus barophilus, the use of oriC diminishes from the lag to the middle of the log phase, and subsequently increases gradually upon entering the stationary phase. Although oriC demonstrates no indispensability, RadA does exhibit essentiality. Notably, a knockdown mutant strain provides confirmation of the pivotal role of RadA in RDR for the first time. Thus, we demonstrate the existence of a tight combination between oriC utilization and homologous recombination to initiate DNA replication along the growth phases. Overall, this study demonstrates how diverse physiological states can influence the initiation of DNA replication, offering insights into how environmental sensing might impact this fundamental mechanism of life. More information: https://pubmed.ncbi.nlm.nih.gov/38421162/ Contact: Jacques OBERTO <jacques.oberto@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
February 19, 2024 10:25 AM
|
EMBO conference "Molecular Biology of Archaea" 23-27th of june 2024 in Palaiseau (near Paris)!
The registration in now open for the next EMBO conference "Molecular Biology of Archaea" that will take place near Paris (Palaiseau) on 23-27th of june 2024! Archaea are fascinating organisms with a bacteria-like morphology, but information-processing systems that are homologous to eukaryotic counterparts, for instance replication, transcription and translation. Recently, novel groups of archaea (e.g. Asgard archaea) have been discovered that represent the closest living relatives to eukaryotes and shed new light on their evolution. Archaeal research has led to seminal change-of-concept mechanistic discoveries in the area of molecular biology, and many studies are currently aiming to unravel the global environmental impact of archaea. Their viruses have a remarkable diversity of morphotypes, exceeding that of bacterial or eukaryotic viruses.The EMBO Workshop on Molecular biology of Archaea will cover cutting-edge research in various fields with a focus on archaeal molecular biology, evolution, and ecology and their interconnections. The proposed talks will present data at different scales ranging from single molecule and structural studies to archaeal physiology, metabolism and ecological impact. This is of high interest as molecular studies of archaea are a very active field, reflecting the wealth of metagenomics information revealing novel biology that can now be addressed by the latest experimental and computational techniques. all information: https://meetings.embo.org/event/24-archaea contact: Tamara BASTA-LE BERRE <tamara.basta@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
February 19, 2024 10:14 AM
|
Transposon sequencing reveals genes enabling insect gut colonization by the symbiont Caballeronia insecticola
High-througput genetic screens with Tn-seq in an insect gut bacterium reveals gut-derived nutrients consumed by the symbiont. Caballeronia insecticola is a bacterium belonging to the Burkholderia sensu lato, able to colonize multiple environments like soils and the gut of the bean bug Riptortus pedestris. We constructed a saturated Himar1 mariner transposon library and revealed by transposon-sequencing (Tn-seq) that 498 protein-coding genes constitute the essential genome of C. insecticola for growth in free-living conditions. By comparing essential gene sets of C. insecticola and seven related Burkholderia s.l. strains, only 120 common genes were identified indicating that a large part of the essential genome is strain-specific. In order to reproduce specific nutritional conditions that are present in the gut of R. pedestris, we grew the mutant library in minimal media supplemented with candidate gut nutrients and identified several condition-dependent fitness-defect genes by Tn-seq. To validate the robustness of the approach, insertion mutants in six fitness genes were constructed and their growth-deficiency in media supplemented with the corresponding nutrient was confirmed. The mutants were further tested for their efficiency in R. pedestris gut colonization, confirming that gluconeogenic carbon sources, taurine and inositol, are nutrients consumed by the symbiont in the gut. Thus, our study provides insights about specific contributions provided by the insect host to the bacterial symbiont. More information: https://doi.org/10.1093/ismeco/ycad001 Contact: Peter MERGAERT <peter.mergaert@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
December 27, 2023 7:57 AM
|
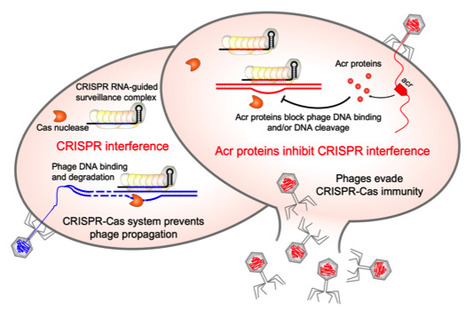
First anti-CRISPR protein that inhibits the CRISPR-Cas defense system in Clostridioides difficile
CRISPR-Cas adaptive immunity defends prokaryotes against their viruses named phages. In response, phages have evolved counter defense strategies to fight back. This study describes the identification of a phage protein to evade CRISPR-Cas immunity in a human pathogen Clostridioides difficile. Clostridioides difficile is the widespread anaerobic spore-forming bacterium that is a major cause of potentially lethal nosocomial infections associated with antibiotic therapy worldwide. Due to the increase in severe forms associated with a strong inflammatory response and higher recurrence rates, a current imperative is to develop synergistic and alternative treatments of C. difficile infections. In particular, phage therapy is regarded as a potential substitute for existing antimicrobial treatments. However, it faces challenges because C. difficile have highly active CRISPR-Cas immunity, which may be a specific adaptation to phage-rich and highly crowded gut environment. To overcome this defense, C. difficile phages must employ anti-CRISPR mechanisms. Here, we present the first anti-CRISPR protein that inhibits the CRISPR-Cas defense system in this pathogen. Our work offers insights into the interactions between C. difficile and its phages, paving the way for future CRISPR-based applications and development of effective phage therapy strategies combined with the engineering of virulent C. difficile infecting phages. More information: doi: 10.1128/msphere.00401-23 Contact: Olga SOUTOURINA <olga.soutourina@i2bc.paris-saclay.fr>
|



 Your new post is loading...
Your new post is loading...