Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
October 6, 3:37 PM
|
New team in the Department of Genome Biology
Cécile Courret, CNRS Researcher and recipient of an ATIP-Avenir grant, has joined the Department of Genome Biology to establish her team ‘Intragenomic Conflict and Evolution’. Cécile's research focus on genetic conflicts and their impact on genome evolution. The basic principles of Mendelian inheritance state that in heterozygous individuals, two alleles have equal chances of being transmitted to the next generation. However, some genetic elements do not follow these rules. These so-called selfish genetic elements, such as transposons or meiotic drivers, bias inheritance in their own favor, often at the expense of the organism. Because they disrupt essential processes like meiosis, heterochromatin regulation, and cell division, selfish elements create a persistent conflict with the host genome. This conflict triggers an evolutionary “arms race,” in which genomes evolve defense mechanisms to counterbalance the harmful effects of these elements. Far from being rare exceptions, such conflicts are now recognized as a major force shaping genome structure and function. My research relies on the Drosophila model and combines genomic, molecular, and cytological approaches to investigate both how selfish elements perturb fundamental biological processes and how organisms respond to mitigate these disruptions. By studying systems where these conflicts are still active in natural populations, as well as the genomic signatures left by past events, Cécile's team aims to better understand how conflicts drive evolutionary innovation and shed light on the molecular basis of essential biological mechanisms. Contact: Cécile Courret, cecile.courret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 29, 2:50 AM
|
Paramecium PolX DNA polymerases keep the genome safe during programmed rearrangements
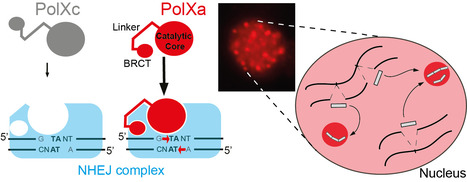
The expansion of PolX DNA polymerases in the ciliate Paramecium tetraurelia has favored the functional diversification of a subset of these enzymes, with enhanced nuclear localization in DNA repair foci during programmed DNA elimination. During its sexual cycle, Paramecium massively rearranges its genome by removing tens of thousands of internal eliminated sequences (IESs) from its developing somatic nucleus. This process involves the introduction of programmed DNA double-strand breaks (DSBs), followed by efficient DSB repair using the non-homologous end joining (NHEJ) pathway. In this system, DSB repair requires not only the core NHEJ factors Ku70/80 and Xrcc4/Lig4, but also additional enzymes that accurately process the 4-base 5′-overhangs of the broken DNA ends created at IES excision sites. In this study, we identify four orthologs of human DNA polymerase lambda in P. tetraurelia —PolXa, PolXb, PolXc, and PolXd— as key players in repairing IES excision junctions. Cells lacking these PolX enzymes accumulate genome-wide DNA damage, including unrepaired double-strand breaks, small deletions, and retained IESs. All four PolX proteins can process DNA ends, but PolXa and PolXb are specifically produced during programmed genome rearrangement and possess a unique internal linker region that enhances their nuclear localization. In contrast to PolXc, we show that PolXa concentrates in nuclear foci along with core NHEJ factors and a Dicer-like enzyme involved in producing IES-specific small RNAs that regulate IES excision. We propose that these foci are the sites where excised IESs are ligated into concatemers, which serve as templates for small RNA production, linking DNA repair to RNA-mediated regulation of genome dynamics. More information: https://academic.oup.com/nar/article/53/7/gkaf286/8113560 Contact: Mireille Bétermier mireille.betermier@i2bc-paris-saclay.fr, Julien Bischerour julien.bischerour@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 11, 5:59 AM
|
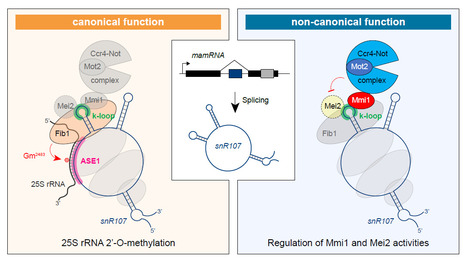
A bifunctional snoRNA guides rRNA 2’-O-methylation and scaffolds gametogenesis effectors
This study uncovers a fission yeast small nucleolar RNA (snoRNA) that guides ribosomal RNA 2’- O-methylation and modulates the activities of RNA-binding proteins involved in gametogenesis, expanding our vision of the non-canonical functions exerted by snoRNAs. Small nucleolar RNAs are non-coding transcripts that guide chemical modifications of RNA substrates and modulate gene expression at the epigenetic and post-transcriptional levels. However, the extent of their regulatory potential and the underlying molecular mechanisms remain poorly understood. In a collaborative work with the I2BC B3S department and NGS facility, the epiRNA-Seq facility in Nancy and the Palancade lab at the Institut Jacques Monod, we have identified a conserved, previously unannotated intronic C/D-box snoRNA, termed snR107, hosted in the fission yeast long non-coding RNA mamRNA and carrying two independent cellular functions. On the one hand, snR107 guides site-specific 25S rRNA 2’-O-methylation and promotes pre-rRNA processing and 60S subunit biogenesis. On the other hand, snR107 associates with the gametogenic RNA-binding proteins Mmi1 and Mei2, mediating their reciprocal inhibition and restricting meiotic gene expression during sexual differentiation. Both functions require distinct cis-motifs within snR107, including a conserved 2’-O-methylation guiding sequence. Together, our results position snR107 as a dual regulator of rRNA modification and gametogenesis effectors, expanding our vision on the non-canonical functions exerted by snoRNAs in cell fate decisions. More Information: https://www.nature.com/articles/s41467-025-58664-y Contact: Mathieu Rougemaille mathieu.rougemaille@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
March 11, 5:05 PM
|
Borders in our chromosomes shape the neighboring domains
Researchers at the I2BC and the Ecole Normale Supérieure found that the borders that create separation between functional domains in our chromosomes have an unexpectedly large influence on those domains themselves. Chromosomes in humans and other mammals are subdivided into “functional neighborhoods” that guide the fidelity of biological processes, including gene regulation and DNA repair. Perturbations of these neighborhoods, resulting in the fusion of adjacent domains, can lead to a variety of diseases, including cancer and developmental disorders.
In a study published in PNAS (Proceedings of the National Academy of Sciences of the USA), scientists from the Noordermeer group at the I2BC, together with their colleagues at the Ecole Normale Supérieure in Paris, report how chromosomal border elements influence the organization of the adjacent functional neighborhoods. By combining genomics-based analysis and biophysical simulations of chromosome structure, they find that the size of these borders is highly diverse, from simple “points” on the chromosome to highly extended zones of transition between neighborhoods. Computer simulations of chromosome behavior of different types of borders in-between revealed an unexpected and far-reaching impact of the borders on their neighboring domains. Rather than creating a static separation, the borders will actively influence the degree of neighborhood mixing due to a previously unrecognized mechanism whereby the borders reel-in the adjacent neighborhoods. At narrow borders, this actively promotes mixing and may cause interference between biological activity on both side of the border. Extended borders buffer against this mechanism by adding additional separation. Border structure can thus constitute a new regulatory layer in the genome to fine-tune biological processes. More information: https://www.pnas.org/doi/10.1073/pnas.2413112122 Contact: Daan Noordermeer daan.noordermeer@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
March 4, 2:57 AM
|
Ribosome profiling and immunopeptidomics reveal tens of novel conserved HIV-1 ORF encoding T cell antigens.
The translatome of HIV-1 reveals tens of alternative open reading frames (ARF), encoding conserved viral antigens. In vivo, ARF-derived peptides elicit potent HIV-specific poly-functional T cell responses mediated by both CD4+ and CD8+ T cells. T lymphocytes play a pivotal role in controlling human immunodeficiency virus type 1 (HIV-1) infection. Their activation relies on the recognition of viral peptides, or antigens, presented on the surface of infected cells by major histocompatibility complex (MHC) molecules. It is widely assumed that these antigens arise solely from canonical HIV proteins. Previously, we showed that HIV-specific T lymphocytes can also target peptides derived from HIV alternative reading frames (ARFs) presented by MHC molecules. However, evidence for ARFs in the HIV genome had thus far been indirect. In this new study, using ribosome profiling (RiboSeq), we identified the complete HIV-1 translatome in infected CD4⁺ T cells. This approach systematically identified virally encoded mRNA sequences actively translated in HIV-infected cells. We found that the HIV-1 genome contains over one hundred ARFs located either in the 5ʹ untranslated region (5ʹUTR) of classical viral genes or overlapping canonical HIV open reading frames. Using two complementary methods: (1) detecting T lymphocytes specific to ARF-derived peptides in PBMCs from people living with HIV and (2) directly isolating ARF-derived peptides bound to MHC molecules via mass spectrometry–based immunopeptidomics—we demonstrate that HIV ARFs encode novel viral antigens capable of eliciting broad and potent T-cell responses. Our results expand the range of HIV antigens that could be harnessed for vaccine development and may also reveal the existence of microproteins or pseudogenes in the HIV genome.
These findings stem from the work of two complementary teams at I2BC: one led by Olivier Namy, a specialist in protein translation, and the other led by Arnaud Moris, an expert in the immune response to HIV. This collaboration also involved French research and clinical teams and the University of Tübingen in Germany. More information: https://doi.org/10.1038/s41467-025-56773-2 Contact: Arnaud Moris arnaud.moris@i2bc.paris-saclay.fr & Olivier Namy olivier.namy@i2bc.paris-saclay.fr Image: Sonhita Chakraborty

|
Scooped by
I2BC Paris-Saclay
January 17, 4:17 AM
|
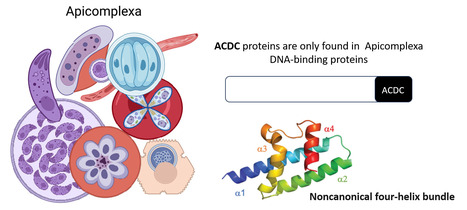
Rocking Science: New ACDC Protein Fold Unveiled
The Apicomplexa-specific ACDC domain adopts a never before seen protein fold. In collaboration with the Nessler team at the I2BC and the Llinás lab at PSU, US, we reveal that the ACDC domain, only found in DNA-binding proteins of the Apicomplexa phylum, grouping several important human pathogens including malaria, has a never before seen protein fold. We also identify potential ligands that may be optimized in the future as protein inhibitors. More information:https://journals.iucr.org/paper?S2059798324012518 Conatct: Joana SANTOS joana.santos@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 9, 2024 3:44 AM
|
Ribosome composition plays a key role in EMT
Ribosome remodeling drives cancer cell transformation: A single protein, RPL36A, triggers EMT. Epithelial-mesenchymal transition (EMT) is a biological process whereby a cell loses its usual characteristics to acquire properties allowing it to move. EMT is involved in embryo formation and tissue repair in adults. However, when altered, EMT contributes to the formation of fibrosis or, in the case of a tumor cell, to the formation of metastases. In cancers, this change is comparable to a cellular metamorphosis that allows cancer cells to become more aggressive and resistant to treatments. EMT is therefore a key player in tumor progression.
EMT is extensively studied at the molecular and cellular levels because a better understanding of this process will enable the development of new therapeutic approaches. However, the translational changes that occur during EMT have so far been very poorly studied.
In this work, we have identified that the overexpression of a single ribosomal protein, called RPL36A, is sufficient to induce EMT. We have also highlighted the various translational changes that occur during EMT.
Our work reveals the importance of ribosome remodeling itself in finely modulating translation during the change in cellular identity that occurs during EMT. More information : https://www.pnas.org/doi/10.1073/pnas.2408114121 ; In French: https://www.insb.cnrs.fr/fr/cnrsinfo/des-modifications-de-la-composition-du-ribosome-accompagnent-les-changements. Contact: Olivier NAMY olivier.namy@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
November 19, 2024 3:27 AM
|
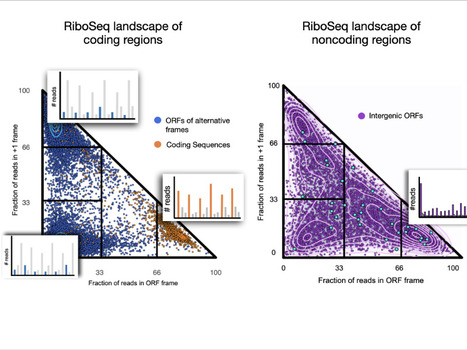
The ribosome profiling landscape of yeast
We explored the ribosome profiling landscape of yeast and showed that noncoding regions are associated with a wide diversity of translation signals among which some led to detectable protein products. Pervasive translation is a widespread phenomenon that plays a critical
role in the emergence of novel microproteins, but the diversity of translation patterns contributing to their generation remains unclear. Based on 54 ribosome profiling (Ribo-Seq) datasets, we investigated the yeast Ribo-Seq landscape using a representation framework that allows the comprehensive inventory and classification of the entire diversity of Ribo-Seq signals, including non-canonical ones. We show that if coding regions occupy specific areas of the Ribo-Seq landscape, noncoding regions encompass a wide diversity of Ribo-Seq signals and, conversely, populate the entire landscape. Our results show that pervasive translation can, nevertheless, be associated with high specificity, with 1055 noncoding ORFs exhibiting canonical Ribo-Seq signals. Using mass spectrometry under standard conditions or proteasome inhibition with an in-house analysis protocol, we report 239 microproteins originating from noncoding ORFs that display canonical but also non-canonical Ribo-Seq signals. Each condition yields dozens of additional microprotein candidates with comparable translation properties, suggesting a larger population of volatile microproteins that are challenging to detect. Our findings suggest that non-canonical translation signals may harbor valuable information and underscore the significance of considering them in proteogenomic studies. Finally, we show that the translation outcome of a noncoding ORF is primarily determined by the initiating codon and the codon distribution in its two alternative frames, rather than features indicative of functionality. Our results enable us to propose a topology of a species’ Ribo-Seq landscape, opening the way to comparative analyses of this translation landscape under different conditions. More information: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03403-7 Contact: Anne Lopes anne.lopes@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
November 3, 2024 12:00 PM
|
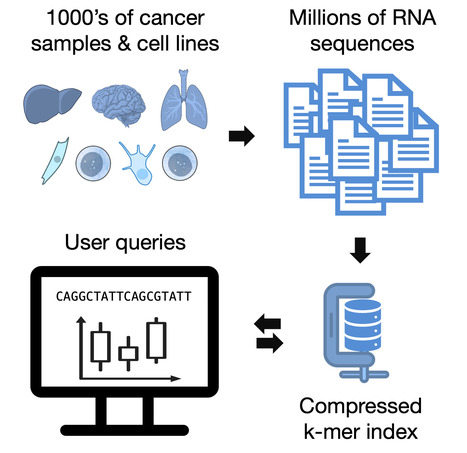
Reference-free exploration of cancer RNA datasets
Bioinformaticians finds new ways to analyze cancer RNA in very large datasets Cancer cells produce an immense diversity of RNA molecules. Using deep sequencing technologies, scientists explore these RNAs to uncover key insights into cancer onset and progression. However, cancer RNA datasets are becoming so large that directly searching them for a given sequence is impossible. Currently, scientists must rely on simplified representations, such as gene expression tables. Yet, many cancer-causing RNAs are absent from standard gene expression tables. The Transipedia consortium, bringing together bioinformaticians from I2BC/University Paris-Saclay, University of Lille, and University of Montpellier, introduces a new approach to mining RNA-seq databases using k-mer indexes. In their latest paper, they demonstrate that an index of 1,019 cancer cell lines can be queried in real time while providing accurate quantification of cancer mutations, fusions, or splicing variants. In future work, the consortium plans to expand this technology to offer access to an even larger database of cancer and normal tissues, aiming to accelerate cancer genomics research. More information: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03413-5 Contact: Daniel Gautheret daniel.gautheret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 29, 2024 5:36 AM
|
Novel, tightly structurally related N-myristoyltransferase inhibitors display equally potent yet distinct inhibitory mechanisms
Peptides fitting the optimal human NMT Gly-myristoylation recognition pattern act as potent inhibitors. Lys-myristoylation-based inhibitors from these peptides were designed. Each series’ inhibitory properties are unique, relying on distinct interactions. N-myristoyltransferases (NMTs) catalyze essential acylations of N-terminal alpha or epsilon amino groups of glycines or lysines. Here, we reveal that peptides tightly fitting the optimal glycine recognition pattern of human NMTs are potent prodrugs relying on a single-turnover mechanism. Sequence scanning of the inhibitory potency of the series closely reflects NMT glycine substrate specificity rules, with the lead inhibitor blocking myristoylation by NMTs of various species. We further redesigned the series based on the recently recognized lysine-myristoylation mechanism by taking advantage of (i) the optimal peptide chassis and (ii) lysine side chain mimicry with unnatural enantiomers. Unlike the lead series, the inhibitory properties of the new compounds rely on the protonated state of the side chain amine, which stabilizes a salt bridge with the catalytic base at the active site. Our study provides the basis for designing first-in-class NMT inhibitors tailored for infectious diseases and alternative active site targeting. More information: https://www-cell-com.insb.bib.cnrs.fr/structure/abstract/S0969-2126(24)00318-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0969212624003186%3Fshowall%3Dtrue Contact: Thierry Meinnel thierry.meinnel@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 17, 2024 5:45 AM
|
De Novo Emerged Gene Search in Eukaryotes with DENSE
DENSE with your favorite genome and uncover its de novo genes! Try our fully automated tool for DE Novo gene SEarch and discover useful metrics to help design your search! The recent discovery of de novo emerged genes, which originate from previously noncoding regions of DNA, challenges traditional views of species evolution. Indeed, the hypothesis of neutrally evolving sequences giving rise to functional proteins is highly unlikely. However, the accumulation of sequencing data revealed that they are widespread, being detected in various eukaryotic species and numerous, with several dozens of examples reported for different organisms. This conundrum has sparked numerous studies to quantify and characterize these genes, aiming to understand their functional roles and contributions to genome evolution. Yet, no fully automated pipeline for their identification is available. Therefore, we introduce DENSE (DE Novo emerged gene SEarch), an automated Nextflow pipeline based on two distinct steps: detection of taxonomically restricted genes (TRGs) through phylostratigraphy, and filtering of TRGs for de novo emerged genes via genome comparisons and synteny search. DENSE is available as a user-friendly command-line tool, while the second step is accessible through a web server upon providing a list of TRGs. Highly flexible, DENSE provides various strategy and parameter combinations, enabling users to adapt to specific configurations or define their own strategy through a rational framework, facilitating protocol communication, and study interoperability. We apply DENSE to seven model organisms, exploring the impact of its strategies and parameters on de novo gene predictions. This thorough analysis across species with different evolutionary rates reveals useful metrics for users to define input datasets, identify favorable/unfavorable conditions for de novo gene detection, and control potential biases in genome annotations. Additionally, predictions made for the seven model organisms are compiled into a requestable database, which we hope will serve as a reference for de novo emerged gene lists generated with specific criteria combinations. More information: https://academic.oup.com/gbe/article/16/8/evae159/7745841 Contact: Anne LOPES anne.lopes@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 23, 2024 10:19 AM
|
Prophage induction can facilitate the in vitro dispersal of multicellular Streptomyces structures
Unveiling a new property of bacteriophage-host interaction in the context of multicellular aggregate dynamics – A multidisciplinary study involving three departments from I2BC (Genomes, Microbiology, and Virology) and a team from MICALIS. Streptomyces are renowned for their prolific production of specialized metabolites with applications in medicine and agriculture. These multicellular bacteria present a sophisticated developmental cycle, and play a key role in soil ecology. Little is known about the impact of Streptomyces-phage on bacterial physiology. In this study, we investigated the conditions governing the expression and production of ‘Samy’, a prophage found in Streptomyces ambofaciens ATCC 23877. This siphoprophage is produced simultaneously with the activation of other mobile genetic elements. Remarkably, the presence and production of Samy increases bacterial dispersal under in vitro stress conditions. Altogether, this study unveiled a new property of a bacteriophage infection in the context of multicellular aggregate dynamics. More information: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3002725 Contact: Stéphanie BURY-MONE stephanie.bury-mone@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 26, 2024 9:11 AM
|
A tumor suppressor that generates subnucleosomes
Researchers from the I2BC, the CNRGH (CEA, National Center of Human Genomics Research), and the University of Edinburgh have revealed how the BAF chromatin remodeler changes the structure and accessibility of genetic material by producing subnucleosomal particles. These findings are published in Nature Structural and Molecular Biology. Mutations in genes encoding subunits of the BAF complex (BRG1/BRM-associated factors, also known as the SWI/SNF complex) are associated with 20% of human cancers, making this complex the second most frequently mutated factor after the p53 tumor suppressor.
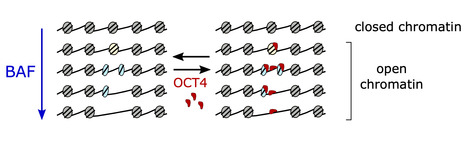
The human genome exists in cells in the form of chromatin, composed of nucleosomes that condense and protect the genetic material. The nucleosomes represent a physical barrier for transcription factors and molecular machinery that regulate gene expression. The BAF complex is known for controlling chromatin opening at gene transcription regulatory elements. Until now, it was believed that the role of the BAF complex was to dissociate the nucleosomes to allow the transcription factors to access DNA.
Using a method that allows them to differentiate histone-free DNA fragments from nucleosomes and histone-containing particles smaller than nucleosomes (subnucleosomes), researchers coordinated by Matthieu Gérard (I2BC) revealed that the BAF complex facilitates the recruitment of the master transcription factor OCT4 using two distinct strategies: i) by generating the well-known histone-free DNA regions, which OCT4 binds if its target DNA motif is present; and ii) by producing a new class of subnucleosomal particles containing 50 to 80 base pairs associated with histones.
They show that the subnucleosomes act as a recruitment platform for OCT4 independently of the presence of its DNA motif. They identify in this publication a molecular mechanism in which the interaction between OCT4 and the subnucleosomes leads to a spectacular expansion of the OCT4 genomic binding interval. This mechanism aims to project OCT4 activity in chromatin opening within a genomic interval up to one order of magnitude larger than the interval bound by OCT4 on histone-free DNA.
This study suggests two main determinants for recruiting transcription factors onto the mammalian genome: the genetic determinant, the transcription factor motif within histone-free DNA, and an epigenetic determinant, which corresponds to the subnucleosomes produced by the BAF complex. More information: https://www.nature.com/articles/s41594-024-01344-0 Contact: Matthieu GERARD matthieu.gerard@i2bc.paris-saclay.fr
|

|
Scooped by
I2BC Paris-Saclay
April 14, 3:19 AM
|
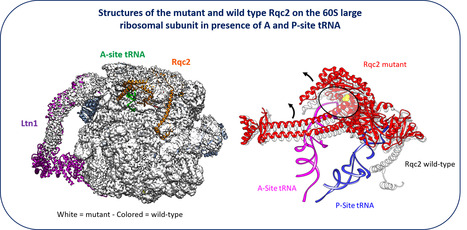
New facet of the Rqc2 protein part of a ribosome rescue pathway in Saccharomyces cerevisiae
The Ribosome Quality Complex protein 2 is a major player in peptide release from stalled ribosomes. Cells employ many quality control mechanisms during protein biosynthesis. Defects impairing messenger RNA decoding can cause ribosomes to stall, thereby distorting gene expression. Ribosome stalling results in ribosomes being sequestered in an inactive state on mRNA and represents a challenge for both ribosome homeostasis and cell survival. Eukaryotic cells prevent the accumulation of potentially toxic aberrant polypeptides and maintain ribosome availability by employing surveillance and clearance mechanisms, including the evolutionarily conserved ribosome-associated quality control complex (RQC). The rescue pathways dissociate the stalled ribosome, leading to the release of a 40S subunit attached to the mRNA and a 60S subunit still attached to a tRNA with the elongating peptide. RQC mediates the ubiquitination, extraction, and rapid degradation of the aberrant nascent peptide. In particular, Rqc2p recruits charged alanyl- and threonyl-tRNAs at the same position as an A-site tRNA, resulting in C-terminal extensions composed of alanine and threonine residues (CAT tails). These CAT tails may expose potential tunnel-embedded lysine residues for ubiquitination by the E3 ligase Ltn1. Using a genetic screen, we identified a mutant allele of RQC2 as involved in peptide release from stalled ribosomes. We characterized the mutant protein both biochemically and structurally, in collaboration with Reynald Gillet’s team (IGDR, Rennes University). We determining how the underlying point mutation reduced tRNA recruitment to the A site, thereby limiting CAT tail formation. These findings provide further mechanistic insight into the role of Rqc2p in ribosome rescue and the maintenance of ribosome homeostasis. More Information:https://pubmed.ncbi.nlm.nih.gov/40187343/ Contact: Céline Fabret celine.fabret@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 7, 5:04 AM
|
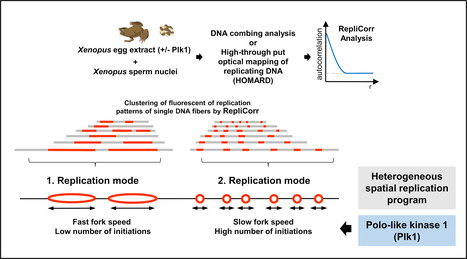
Yin-Yang during DNA replication
DNA duplication of the genetic material is a crucial step of cell proliferation. The study reveals that this step comprises two opposed strategies, coordinated by the enzyme Plk1. In vertebrates, DNA replication involves the activation of thousands of starting points for DNA copying, known as “origins of replication”. These points are irregularly scattered across the genome. However, the precise mechanisms controlling the coordinated activation of these starting points and the regulation of the rate at which DNA is copied remain poorly understood. Any dysfunction can lead to genetic abnormalities and promote the development of cancers.
The scientists studied DNA replication using cell-free extracts from the eggs of the amphibian Xenopus laevis. This model is quite similar to replication in human cells, and has made it possible to develop a new method for analyzing the distribution of replication starting points (initiations) and the speed of replication in individualized DNA molecules. Combining this analysis with kinetic models, the scientists discovered that DNA replication relies on two dynamic and opposing strategies:
A fast strategy: high replication speed, but with fewer starting points.
A slow strategy: a slower replication speed compensated by the activation of numerous starting points.
In this context, the scientists determined that the enzyme Polo-like kinase 1 (Plk1) played a key role in coordinating these two strategies. Plk1, frequently mutated in many cancers, regulates this balance, opening up new perspectives on the control of replication and its implications in oncology. (published by Paris-Saclay and CNRS Biologie Actu (https://www.insb.cnrs.fr/fr/cnrsinfo/yin-yang-dans-la-duplication-des-chromosomes ) More Information: https://doi.org/10.1093/nar/gkaf007 Contact: Arach Goldar arach.goldar@i2bc.paris-saclay.fr Kathrin Marheineke kathrin.marheineke@ijm.fr

|
Scooped by
I2BC Paris-Saclay
March 4, 3:20 AM
|
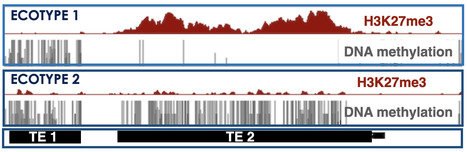
Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3
This study sheds light on an alternative mode of Transposable Element silencing associated with H3K27me3 instead of DNA methylation in flowering plants; it also indicates dynamic switching between the two epigenetic marks at the species level, a new paradigm that might extend to other multicellular eukaryotes. The DNA/H3K9 methylation and Polycomb-group proteins (PcG)-H3K27me3 silencing pathways have long been considered mutually exclusive and specific to transposable elements (TEs) and genes, respectively in mammals, plants, and fungi. However, H3K27me3 can be recruited to many TEs in the absence of DNA/H3K9 methylation machinery and sometimes also co-occur with DNA methylation.
In this study, the team of Angélique Déléris showed that TEs can also be solely targeted and silenced by H3K27me3 in wild-type Arabidopsis plants. These H3K27me3-marked TEs not only comprise degenerated relics but also seemingly intact copies that display the epigenetic features of responsive PcG target genes as well as an active H3K27me3 regulation. The authors also showed that H3K27me3 can be deposited on newly inserted transgenic TE sequences in a TE-specific manner indicating that silencing is determined in cis. Finally, a comparison of Arabidopsis natural accessions reveals the existence of a category of TEs—which we refer to as “bifrons”—that are marked by DNA methylation or H3K27me3 depending on the accession. This variation can be linked to intrinsic TE features and to trans-acting factors and reveals a change in epigenetic status across the TE lifespan.
Overall, this study shedded light on an alternative mode of TE silencing associated with H3K27me3 instead of DNA methylation in flowering plants. It also suggests dynamic switching between the two epigenetic marks at the species level, a new paradigm that might extend to other multicellular eukaryotes. More Information: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03466-6 Contact: Angélique Déléris angelique.deleris@i2bc.apris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
January 30, 3:50 AM
|
Genetic differentiation in the MAT-proximal region is not sufficient for suppressing recombination in Podospora anserina
A large genomic region in Podospora anserina remains entirely devoid of crossovers, despite being fully colinear. Recombination is advantageous over the long-term, as it allows efficient selection and purging deleterious mutations. Nevertheless, recombination suppression has repeatedly evolved in sex chromosomes. In fungi, sexual compatibility is driven by a locus called mating-type (mat), around which recombination suppression is also often observed.
The evolutionary causes for recombination suppression and the proximal mechanisms preventing crossing overs are poorly understood. Several hypotheses have recently been suggested based on theoretical models, and in particular, that divergence could accumulate neutrally around a sex-determining region and reduce recombination rates, a self-reinforcing process that could foster progressive extension of recombination suppression.
We used the ascomycete fungus Podospora anserina for investigating these questions: a 0.8 Mbp region around its mat locus is non-recombining (called MAT-proximal region), despite being collinear between the two mating types. This fungus is mostly selfing, resulting in highly homozygous individuals, except in the non-recombining region around the mating-type locus that displays differentiation between mating types. Here, we test the hypothesis that sequence divergence alone is responsible for recombination cessation. We replaced one mat idiomorph by the sequence of the other, to obtain compatible strains isogenic in the MAT-proximal region. Crosses showed that recombination was still suppressed in that context, indicating that other proximal mechanisms than inversions or mere sequence divergence are responsible for recombination suppression in this fungus.
This finding suggests that selective mechanisms likely acted for suppressing recombination, or the spread of epigenetic marks, as the neutral model based on mere nucleotide divergence does not seem to hold in P. anserina. More information: https://doi.org/10.1093/g3journal/jkaf015 Contact: Pierre GROGNET pierre.grognet@universite-paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
December 16, 2024 5:49 AM
|
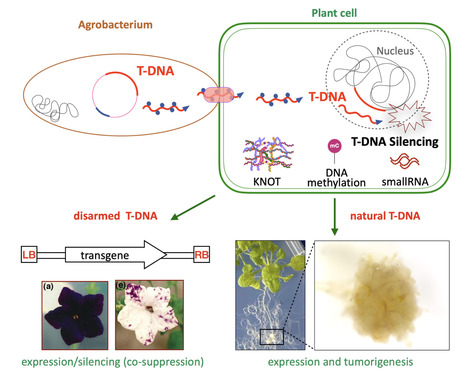
Epigenetic control of T-DNA during transgenesis and pathogenesis
T-DNAs are mobile elements transferred from pathogenic Agrobacterium to plants that reprogram host cells into hairy roots or tumors, and which are used as disarmed forms to deliver transgenes in plants. Here we review the mechanisms that silence the expression of T-DNAs in transgenic plants as well as during pathogenesis. Mobile elements known as T-DNAs are transferred from pathogenic Agrobacterium to plants and reprogram the host cell to form hairy roots or tumors. Disarmed nononcogenic T-DNAs are extensively used to deliver transgenes in plant genetic engineering. Such T-DNAs were the first known targets of RNA silencing mechanisms, which detect foreign RNA in plant cells and produce small RNAs that induce transcript degradation. These T-DNAs can also be transcriptionally silenced by the deposition of epigenetic marks such as DNA methylation and the dimethylation of lysine 9 (H3K9me2) in plants. Here, we review the targeting and the roles of RNA silencing and DNA methylation on T-DNAs in transgenic plants as well as during pathogenesis. In addition, we discuss the crosstalk between T-DNAs and genome-wide changes in DNA methylation during pathogenesis. We also cover recently discovered regulatory phenomena, such as T-DNA suppression and RNA silencing-independent and epigenetic-independent mechanisms that can silence T-DNAs. Finally, we discuss the implications of findings on T-DNA silencing for the improvement of plant genetic engineering. More information: https://academic-oup-com.insb.bib.cnrs.fr/plphys/advance-article/doi/10.1093/plphys/kiae583/7876130 Contact: Angélique DELERIS angelique.deleris@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
November 19, 2024 3:50 AM
|
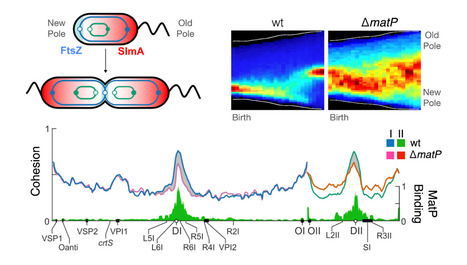
MatP local enrichment delays segregation independently of tetramer formation and septal anchoring in Vibrio cholerae
The study of Vibrio cholerae MatP reveals the degree of integration of the plasmid-derived chromosome carried by the bacterium, and suggests that the ancestral role of MatP is to promote cell division at the end of the chromosome replication and segregation cycle. V. cholerae harbours a primary chromosome derived from the monochromosomal ancestor of the Vibrionales (ChrI) and a secondary chromosome derived from a megaplasmid (ChrII). The coordinated segregation of the replication terminus of both chromosomes (TerI and TerII) determines when and where cell division occurs. ChrI encodes a homolog of Escherichia coli MatP, a protein that binds to a DNA motif (matS) that is overrepresented in replication termini. Here, we use a combination of deep sequencing and fluorescence microscopy techniques to show that V. cholerae MatP structures TerI and TerII into macrodomains, targets them to mid-cell during replication, and delays their segregation, thus supporting that ChrII behaves as a bona fide chromosome. We further show that the extent of the segregation delay mediated by MatP depends on the number and local density of matS sites, and is independent of its assembly into tetramers and any interaction with the divisome, in contrast to what has been previously observed in E. coli. More information: https://www.nature.com/articles/s41467-024-54195-0 Contact: E. Galli elisa.galli@i2bc.paris-saclay.fr & FX Barre francois-xavier.barre@i2bc.paris-saclay.fr
- Are you interested in Parasitology? or - Would you like to discover a field of research at the interface between Biology and Chemistry? or - Would you like to discover new research methods or study models? Come and attend the Workshop: Practical information: - Date: 11/27/2024
- Times: 9.15 a.m. to 12.15 p.m. (detailed program above)
- Address: Bâtiment Henri Moissan, 17, avenue des Sciences, 91400 ORSAY, France
- Room: 2000 (HM1 building)
- Program and theme (details attached): 3 parasitology specialists come to talk about their latest research findings on malaria and leishmaniasis. Come and listen to their lectures and have lunch with them.
- Registration (deadline 11/20/2024): registration is free but compulsory by clicking on this link
- Module validation: This seminar could validate a doctoral school module for doctoral students.
Via Life Sciences UPSaclay

|
Scooped by
I2BC Paris-Saclay
October 30, 2024 8:53 AM
|
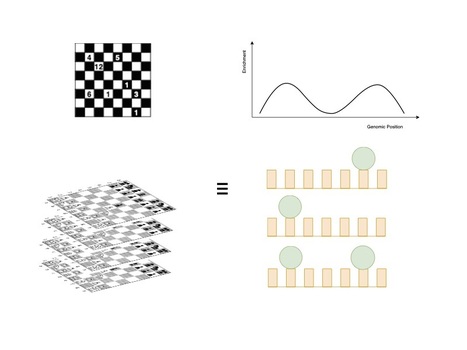
The next-generation sequencing—chess problem
By comparing the collection of Next-Generation Sequencing data to the superimposition of several chess games, we demonstrate the limits of current temporal analysis approaches. The development of next-generation sequencing (NGS) technologies paved the way for studying the spatiotemporal coordination of cellular processes along the genome. However, data sets are commonly limited to a few time points, and missing information needs to be interpolated. Most models assume that the studied dynamics are similar between individual cells, so that a homogeneous cell culture can be represented by a population-wide average. Here, we demonstrate that this understanding can be inappropriate. We developed a thought experiment—which we call the NGS chess problem—in which we compare the temporal sequencing data analysis to observing a superimposed picture of many independent games of chess at a time. The analysis of the spatiotemporal kinetics advocates for a new methodology that considers DNA-particle interactions in each cell independently even for a homogeneous cell population. More information: https://academic.oup.com/nargab/article/6/4/lqae144/7833696 Contact: Julie Soutourina julie.soutourina@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 17, 2024 5:56 AM
|
DciA secures bidirectional replication initiation in Vibrio cholerae
DciA secures bidirectional replication initiation in Vibrio cholerae. Replication is initiated bidirectionally in the three domains of life by the assembly of two replication forks at an origin of replication. This is made possible by the recruitment of two replicative helicases to a nucleoprotein platform built at the origin of replication with the initiator protein. The reason why replication is initiated bidirectionally has never been experimentally addressed due to the lack of a suitable biological system. Using genetic and genomic approaches, we show that upon depletion of DciA, replication is no longer initiated bidirectionally at the origin of replication of Vibrio cholerae chromosome 1. We show that following unidirectional replication on the left replichore, nascent DNA strands at ori1 anneal to each other to form a double-stranded DNA end. While this DNA end can be efficiently resected in recB+ cells, only a few cells use it to trigger replication on the right replichore. In most DciA-depleted cells, chromosome 1 is degraded leading to cell death. Our results suggest that DciA is essential to ensuring bidirectional initiation of replication in bacteria, preventing a cascade of deleterious events following unidirectional replication initiation. More information: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae795/7759985 Contact: Amelie BESOMBES amelie.besombes@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
October 1, 2024 6:27 AM
|
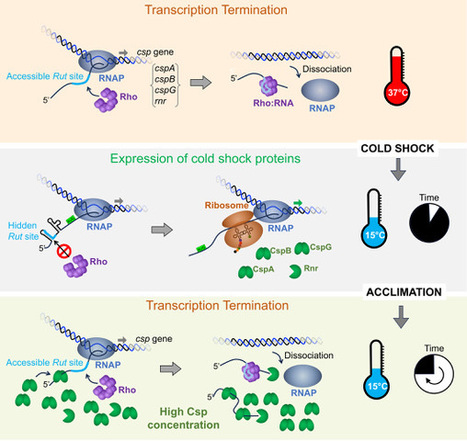
Rho-dependent transcriptional switches regulate the bacterial response to cold shock
How enteric bacteria handle a cold snap Bacteria that inhabit warm-blooded hosts, particularly enteric pathogens and commensals, often spend prolonged periods outside their hosts in cooler environments. The transition from a warm to a cold external environment typically occurs abruptly when bacteria are released from the host. Understanding how these microorganisms rapidly adapt and thrive at low temperatures is crucial for defining their persistence on contaminated surfaces and their spread to humans and animals.
At the molecular level, the primary effect of cold shock on bacterial physiology is the inhibition of protein synthesis, largely due to increased RNA structuring. Bacteria respond by synthesizing a set of RNA chaperone proteins that either unfold RNA secondary structures or degrade misfolded, untranslatable RNA. The synthesis of these “cold-shock response” (CSR) proteins is driven by cold-induced secondary structures in the 5’ untranslated regions of their mRNAs, which expose ribosome binding sites and facilitate translation initiation. As CSR proteins accumulate, their RNA unfolding activity on their own mRNAs acts as a negative feedback mechanism to reduce synthesis once bacteria acclimate to low temperatures.
In a study published in Molecular Cell, researchers from the I2BC, in collaboration with a group from the CBM, CNRS, Orleans, demonstrated that the very RNA structures that activate CSP mRNA translation act earlier to prevent premature transcription termination by sequestering the binding sites for the termination factor Rho. Remarkably, these conditional terminators function as dual-input, antagonistic riboswitches that regulate Rho access to mRNA based on temperature and CSR protein concentration. Together with established post-transcriptional mechanisms, this allows tight and timely control of CSR gene expression: an initial burst during cold shock followed by attenuation during acclimation. This work establishes a novel paradigm for how RNA thermosensors modulate gene expression. more information: https://www.sciencedirect.com/science/article/pii/S1097276524006324 Contact: nara.figueroa@i2bc.paris-saclay.fr lionello.bossi@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 23, 2024 10:17 AM
|
TLN468 changes the pattern of tRNA used to read through premature termination codons in CFTR
TLN468 promotes PTC readthrough by preferentially incorporating specific tRNAs based on the mutation. Nonsense mutations account for 12 % of cystic fibrosis (CF) cases. The presence of a premature termination codon (PTC) leads to gene inactivation, which can be countered by the use of drugs stimulating PTC readthrough, restoring production of the full-length protein. We recently identified a new readthrough inducer, TLN468, more efficient than gentamicin.
We measured the readthrough induced by these two drugs with different cystic fibrosis transmembrane conductance regulator (CFTR) PTCs. We then determined the amino acids inserted at the S1196X, G542X, W846X and E1417X PTCs of CFTR during readthrough induced by gentamicin or TLN468. TLN468 significantly promoted the incorporation of one specific amino acid, whereas gentamicin did not greatly modify the pro- portions of the various amino acids incorporated relative to basal conditions. The function of the engineered missense CFTR channels corresponding to these four PTCs was assessed with and without potentiator. For the recoded CFTR, except for E1417Q and G542W, the PTC readthrough induced by TLN468 allowed the expression of CFTR variants that were correctly processed and had significant activity that was enhanced by CFTR modu- lators. These results suggest that it would be relevant to assess the therapeutic benefit of TLN468 PTC sup- pression in combination with CFTR modulators in preclinical assays. More information: https://www.cysticfibrosisjournal.com/article/S1569-1993(24)00802-6/fulltext Contact: Olivier NAMY olivier.namy@i2bc.paris-saclay.fr
|




 Your new post is loading...
Your new post is loading...