Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
March 1, 2022 5:06 AM
|
HubP-dependent cell pole organization in Vibrio cholerae
By comparative proteomics we identified several novel proteins localize to the cell pole in Vibrio cholerae. Characterization of these proteins revealed an ever more versatile role for the landmark cell pole organizer HubP and identify novel mechanisms of motility regulation. Cell polarity is the result of controlled asymmetric distribution of protein macrocomplexes, genetic material, membrane lipids and cellular metabolites, and can play crucial physiological roles not only in multicellular organisms but also in unicellular bacteria. In the opportunistic cholera pathogen Vibrio cholerae, the polar landmark protein HubP tethers key actors in chromosome segregation, chemotaxis and flagellar biosynthesis and thus converts the cell pole into an important functional microdomain for cell proliferation, environmental sensing and adaptation between free-living and pathogenic life-styles. Using a comparative proteomics approach, we here-in present a comprehensive analysis of HubP-dependent cell pole protein sorting and identify novel HubP partners including ones likely involved in cell wall remodeling (DacB), chemotaxis (HlyB) and motility regulation (MotV and MotW). Unlike previous studies which have identified early roles for HubP in flagellar assembly, functional, genetic and phylogenetic analyses of its MotV and MotW partners suggest a direct role in flagellar rotary mechanics and provide new insights into the coevolution and functional interdependence of chemotactic signaling, bacterial motility and biofilm formation. This work was done in collaboration of Petya Krasteva team, former in B3S Dept of I2BC currently in IECB Bordeaux. More information: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009991 Contact: Yoshi Yamaichi <yoshiharu.yamaichi@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
February 21, 2022 8:54 AM
|
STÉPHANIE BURY MONÉ: REVEALING THE HIDDEN POTENTIAL OF BACTERIA

|
Scooped by
I2BC Paris-Saclay
December 20, 2021 3:44 AM
|
Intergenic ORFs as elementary structural modules of de novo gene birth and protein evolution
Intergenic ORFs of S. cerevisiae encode the elementary building bricks of protein structures and can provide the raw material for de novo gene birth and protein evolution. The noncoding genome plays an important role in de novo gene birth and in the emergence of genetic novelty. Nevertheless, how noncoding sequences’ properties could promote the birth of novel genes and shape the evolution and the structural diversity of proteins remains unclear. In this work, in collaboration with Namy's team, IMPMC and DSIMB lab, we show that the Intergenic ORFs (Open Reading Frames) of S. cerevisiae encode the elementary building bricks of protein structures and can provide the raw material for de novo gene birth and protein evolution. In particular, we show that the noncoding genome contain a vast amount of Intergenic ORFs encoding foldable peptides. The latter can serve as starting points for de novo gene emergence or be integrated into pre-existing proteins, thus contributing to protein modularity and participating in protein evolution. Then, we investigated the early stages of de novo gene birth by reconstructing the ancestral sequences of 70 yeast de novo genes and characterized the sequence and structural properties of intergenic ORFs with a strong translation signal. This enabled us to highlight sequence and structural factors determining de novo gene emergence. In particular, we showed that ancestral intergenic ORFs and highly translated intergenic ORFs are enriched in ORFs encoding peptides with a strong folding potential, thereby giving a central role to protein foldability in the emergence of new genes. Finally, we showed a strong correlation between the fold potential of de novo proteins and one of their ancestral amino acid sequences, reflecting the intimate relationship between the noncoding genome and the protein structure universe.

|
Scooped by
I2BC Paris-Saclay
October 14, 2021 2:59 AM
|
BitQT: a graph-based approach to the quality threshold clustering of molecular dynamics
A fast and yet accurate QT clustering method: BitQT, example of application to MD trajectories The term clustering designates a comprehensive family of unsupervised learning methods, allowing to group similar elements into sets called clusters. The Quality Threshold (QT) clustering algorithm stands out as an ideal option whenever highly correlated elements are needed to be returned as clusters. QT guarantees that all cluster members will maintain a collective similarity established by a user-defined threshold. Unfortunately, the high computational cost of this algorithm for processing big data limits its application domain. In this work, we proposed a methodological parallel between QT clustering and another well-known algorithm in Graph Theory, the Maximum Clique Problem. We targeted Molecular Dynamics (MD) trajectories as an object of study, by representing molecular conformations as nodes of a graph whose edges imply a mutual similarity between conformations. Using a binary-encoded similarity matrix coupled to the exploitation of bitwise operations to extract clusters significantly contributed to reaching a very affordable algorithm compared to the few implementations of QT for MD available in the literature. Our alternative provides results in good agreement with the exact one, while strictly preserving the collective similarity of clusters. We believe methodological parallels discussed here may be translated to other areas where the QT hallmarks are helpful (e.g. transcriptomics). More details here Contact person: Fabrice Leclerc Follow the team

|
Scooped by
I2BC Paris-Saclay
October 14, 2021 2:36 AM
|
Role of polycomb in the control of transposable elements
This opinion piece reviews for the first time the evidences across kingdoms that Polycomb (PcG) Repressive Complex 2, thought to be dedicated to the epigenetic silencing of protein-coding genes, can also target, and even silence, transposable elements: could an ancestral role of PcG proteins be to silence transposable elements? It is generally considered that Polycomb Repressive Complex 2 deposits the histone mark H3K27me3 on silent protein coding genes, while transposable elements are repressed by DNA and/or H3K9 methylation. Yet, there is increasing evidence that the Polycomb repressive complexes also target and even silence transposable elements in representatives of several distantly related eukaryotic lineages. In plants and animals, H3K27me3 is present on transposable elements in mutants and specific cell types devoid of DNA methylation. In this opinion, we summarize the experimental evidence for this phenomenon across the eukaryotic kingdom, and discuss its functional and evolutionary significance. We hypothesize that an ancestral role of Polycomb group proteins was to silence transposable elements. More details here Contact person: Angélique Déléris Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 5:37 AM
|
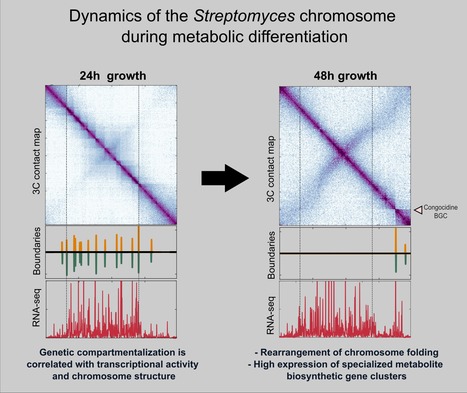
The folding dynamics of Streptomyces' chromosome during metabolic differentiation is revealed
A joint collaborative effort of the Microbiology, Genome Biology Departments and the Sequencing Facility of the I2BC unveiled the dynamics of the linear chromosome of Streptomyces ambofaciens, during metabolic differentiation. Streptomyces are soil bacteria mostly known for their complex life cycle (uni- to multi-cellular transition, sporulation, metabolic differentiation) and their prolific specialized metabolism (e.g. antibiotics and pigments), widely exploited in medicine, agriculture and the food industry. Their genome possesses remarkable characteristics including a large size (6-12 Mb) and a high GC content (circa 72%) and, even more unusual in bacteria, a linear configuration. Interestingly, Streptomyces chromosomes present a remarkable genetic compartmentalization, with a distinguishable central region harboring core genes and two terminal regions enriched in specialized metabolite biosynthetic gene clusters. The molecular mechanisms governing the structure and function of these compartmentalized chromosomes remain mostly unknown. In this work, teams of the Genome Biology, the Microbiology Department and the Sequencing Facility of the I2BC in collaboration with the DynAMIC (Lorraine University -INRAe) and the TIMC-IMAG ( Grenoble Alpes University - CNRS) Institutes, show that chromosome structure in Streptomyces ambofaciens correlates with genetic compartmentalization during exponential phase. Conserved, large and highly transcribed genes form boundaries that segment the central part of the chromosome into domains, whereas the terminal ends tend to be transcriptionally quiescent compartments with different structural features. The onset of metabolic differentiation is accompanied by a rearrangement of the chromosome architecture, from a rather ‘open’ to a ‘closed’ conformation, in which highly expressed specialized metabolite biosynthetic genes form new domain boundaries. Altogether, these results indicate that the linear chromosome of S. ambofaciens is partitioned into structurally distinct entities, suggesting a link between chromosome folding, gene expression and genome evolution. More details here Contact persons: Virginia Lioy & Stephanie Bury-Mone Follow the team
|

|
Scooped by
I2BC Paris-Saclay
February 21, 2022 9:09 AM
|
New member has joined the Programmed Genome Rearrangement Team in I2BC Genome Biology Department.
My name is Valerio VITALI, I am a new Italian postdoc in the Programmed Genome Rearrangement Team (Bétermier Lab) since Feb 1st. Under the guidance of Mireille—and in close collaboration with the team—I am tasked to design and implement creative strategies to characterize the complex machinery that catalyzes developmentally programmed DNA rearrangements in Paramecium (removal of intervening sequences coupled with DNA repair).
I have received my PhD in Evolutionary Genomics and Bioinformatics from the University of Münster (WWU), Germany. While at the Institute for Evolution and Biodiversity, my research has focused on the highly plastic genome architecture of Paramecium, and its evolutionary potential. During my doctoral and (brief) post-doctoral research, I have gained transferable experience in comparative genomics and trancriptomics, single cell whole-genome sequencing, and the analysis of Structural Variants (SVs) from NGS data.
When not in the Lab, I enjoy running, reading books about evolution and aging, learning new languages (or at least trying!) and cooking traditional meals.
I am excited to have joined the Genome Biology Department at the I2BC and can't wait to greatly expand my knowledge of Genome and Cell Biology, Biochemistry, Biophysics, Structural biology.....and beyond. GitHub Profile: https://github.com/biowalter/senes Full list of Publications: https://scholar.google.com/citations?hl=en&user=BI3IdwwAAAAJ

|
Scooped by
I2BC Paris-Saclay
February 21, 2022 8:41 AM
|
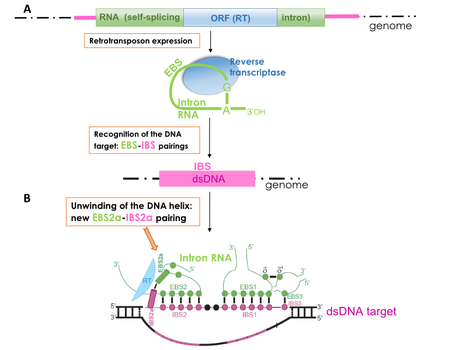
A new RNA–DNA interaction required for integration of group II intron retrotransposons into DNA targets
A novel structural device used by bacterial retrotransposons to efficiently invade host genomes was identified. These results will also contribute to the rational design of new RNA-based gene targeting systems for bacterial genome engineering. Mobile group II introns are site-specific retrotransposable elements widespread in bacterial genomes and in organelles (mitochondria and chloroplasts) of plants, algae and fungi. They constitute the most abundant class of retrotransposons in bacteria and play a major role in the diversification and evolution of bacterial genomes. Mobile group II introns are composed of a large and highly structured self-splicing intron and an intron-encoded reverse transcriptase. They colonize genomes by 'Retrohoming', a very efficient and specific pathway that is operated by the combined activities of the self-splicing intron and its reverse transcriptase. During the first stage of mobility, the intron RNA catalyzes its own insertion directly into the DNA target site. Recognition of the proper target rests primarily on multiple base pairing interactions between the intron segments ‘EBS’ and complementary 'IBS' sequences present on the double-stranded DNA target (Figure 1A). Remarkably, the sequence of the EBS sites can be modified, thus allowing to 'reprogram' a group II intron to specifically integrate into a desired DNA target. This feature is at the basis of the powerful 'Targetron' technology, now commonly used for bacterial genome engineering. Using bioinformatics and genetic approaches, Maria Costa and collaborators have identified a novel base pairing interaction between the intron RNA and the DNA target that is essential for intron mobility. This pairing, named 'EBS2a-IBS2a', adopts a Watson–Crick geometry and helps inducing a structural conformation that favors the opening of the DNA double helix and the integration of the retrotransposon into the genome (Figure 1B). The authors also showed that the EBS2a-IBS2a interaction is perfectly 'reprogrammable'. This work has direct biotechnological applications by allowing the development of new 'targetrons' capable of inserting themselves into genomic sites that were previously inaccessible to disruption by these systems. More information here. Contact person: Maria Costa

|
Scooped by
I2BC Paris-Saclay
November 16, 2021 4:22 AM
|
A Gold medal and the Best Inclusivity Prize for the iGEM GO Paris-Saclay team 2021 supported by I2BC!
The GO Paris-Saclay 2021 team won a gold medal and the "Best Inclusivity" prize at the iGEM 2021 international synthetic biology competition on Sunday, November 14. The team's project "EndoSeek" aims to develop a new diagnostic tool for endometriosis, which is a painful and poorly understood pathology caused by the proliferation of uterine cells outside the uterus. Worldwide, this disease affects about 10% of women and can take up to 7 years to diagnose. Based on preliminary studies using small patient cohorts, certain blood microRNAs (miRNAs) may be biomarkers of the disease.
The team has developed a machine learning program that will allow identification of new endometriosis biomarkers with future cohorts. In experiments conducted this summer at I2BC, students exploited Cas13a and Cas14a1 nucleases for miRNA detection. They created a video game to educate adults and children over the age of 10 about endometriosis and synthetic biology. Finally, following their dialogue with patients and physicians, they questioned the ethical implications of diagnostics.
The team was awarded the Best Inclusivity Award for outstanding efforts to include people with diverse identities. The students thought about the position of LGBT+ people in their project and created a multi-language, voice-assisted website with color customization.
This project was supported by the I2BC, the Faculty of Sciences of the University of Paris-Saclay, La Diagonale Paris-Saclay, EUGLOH (European University Alliance for Global Health), the Graduate School Life Sciences and Health, IDT and Promega. Team 2021 supervisors included Téo Hébra (ICSN) and 5 members of the I2BC: Philippe Bouloc, Stéphanie Bury-Moné, Emma Piattelli, Ombeline Rossier and Charlène Valadon.
For more information, see the wiki https://2021.igem.org/Team:GO_Paris-Saclay and the video https://video.igem.org/w/ihqYR3UfveimEtZVv7x6jE.

|
Scooped by
I2BC Paris-Saclay
October 14, 2021 2:45 AM
|
Characterization of the Radiation Desiccation Response Regulon of the Radioresistant Bacterium Deinococcus radiodurans by Integrative Genomic Analyses
Characterization of the Radiation Desiccation Response
Regulon of the Radioresistant Bacterium Deinococcus
radiodurans by Integrative Genomic Analyses Numerous genes are overexpressed in the radioresistant bacterium Deinococcus radiodurans after exposure to radiation or prolonged desiccation. It was shown that the DdrO and IrrE proteins play a major role in regulating the expression of approximately twenty genes. At present, many questions remain, such as the number of genes regulated by the DdrO regulator. Here, we present the first ChIP-seq analysis performed at the genome level in Deinococcus species coupled with RNA-seq, which was achieved in the presence or not of DdrO. We also resequenced our laboratory stock strain of D. radiodurans R1 ATCC 13939 to obtain an accurate reference for read alignments and gene expression quantifications. We highlighted genes that are directly under the control of this transcriptional repressor and showed that the DdrO regulon in D. radiodurans includes numerous other genes than those previously described, including DNA and RNA metabolism proteins. These results thus pave the way to better understand the radioresistance pathways encoded by this bacterium and to compare the stress-induced responses mediated by this pair of proteins in diverse bacteria. More details here Contact person: Fabrice Confalonieri Follow the team

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 6:04 AM
|
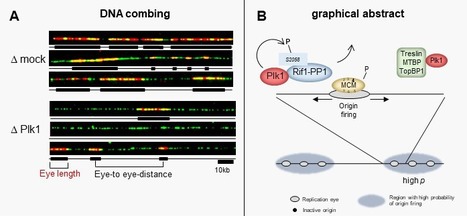
Polo-like kinase 1 (Plk1) regulates replication origin activation and interacts with Rif1
Using different approaches, the group of K. Marheineke in collaboration with A. Goldar (J. Soutourina group) and the proteomics platforms at I2CB and at Necker hospital found a new mechanism of how the kinase Plk1 promotes DNA replication via inhibiting the replication repressor Rif1. The activation of eukaryotic DNA replication origins needs to be strictly controlled at multiple steps in order to faithfully duplicate the genome and to maintain its stability. How the checkpoint recovery and adaptation protein Polo-like kinase 1 (Plk1) regulates the firing of replication origins during non-challenged S phase remained an open question. Using DNA fiber analysis, the group of Kathrin Marheineke in collaboration with Arach Goldar (J. Soutourina lab) show that immunodepletion of Plk1 in the Xenopus in vitro system decreases replication fork density and initiation frequency. Numerical analyses suggest that Plk1 reduces the overall probability and synchrony of origin firing. We used quantitative chromatin proteomics and co-immunoprecipitations in collaboration with SiCAPS I2BC proteomics platform to demonstrate that Plk1 interacts with firing factors MTBP/Treslin/TopBP1 as well as with Rif1, a known regulator of replication timing. Phosphopeptide analysis by LC/MS/MS show that the C-terminal domain of Rif1, which is necessary for its repressive action on origins through protein phosphatase 1 (PP1), can be phosphorylated in vitro by Plk1 on S2058 in its PP1 binding site. The phosphomimetic S2058D mutant interrupts the Rif1-PP1 interaction and modulates DNA replication. Collectively, our study provides molecular insights into how Plk1 regulates the spatio-temporal replication program and suggests that Plk1 controls origin activation at the level of large chromatin domains in vertebrates. More details here Contact persons: Kathrin Marheineke & Arach Goldar Follow the teams of Kathrin Marheineke and of Arach Goldar

|
Scooped by
I2BC Paris-Saclay
September 16, 2021 5:18 AM
|
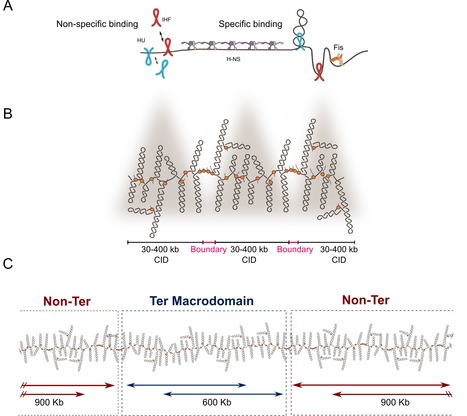
Revisiting the multiscale structuring of bacterial chromosomes
Do you know how bacteria organize their chromosomes allowing key biological processes for the cell? Here we review old and new factors that are essential for the structuring of the bacterial chromosome. During the last decades, works have shown that bacterial chromosomes are not just molecules of DNA inside a bag composed of lipids, peptidoglycan and proteins. Recent advances have not only confirm the multiscale organization of bacterial chromosome, but also have added new layers on chromosome organization. Briefly, DNA supercoiling is organized into stochastic 10-kb domains included in larger 40–300-kb chromosomal interaction domains (CIDs) delimited by long and highly expressed gene clusters. The bacterial chromatin composed of multiple nucleoid associated proteins (NAPs) bound to DNA not only influences gene expression but also modulates, together with bacterial SMC complexes, the folding and disposition of the chromosome in the cell. Although the main structural layers and their key organizers start to emerge, the complete picture of chromosome organization in bacteria remains elusive. In this article, researchers of the Genome Biology Department of the I2BC review not only the classical well-known factors involved in chromosome organization but also novel components that have recently been shown to dynamically shape the 3D structuring of the bacterial genome. More details here Contact person: Frédéric Boccard Follow the team
|




 Your new post is loading...
Your new post is loading...