Your new post is loading...

|
Scooped by
Dr. Stefan Gruenwald
August 18, 2023 8:50 PM
|
The poisonous birds inhabit one of Earth’s most pristine rainforests, a place as exotic as no other in the world. Hearing the words poisonous and bird coupled will be an eye-opener for most. But poisonous birds actually exist. And now, more species have been discovered in New Guinea’s jungles.
"We managed to identify two new species of poisonous birds on our most recent trip. These birds contain a neurotoxin that they can both tolerate and store in their feathers," says Knud Jønsson of the Natural History Museum of Denmark.
Jønsson and fellow UCPH researcher, Kasun Bodawatta have been on an Indiana Jones-like research trip, risking life and limb to regularly warring tribespeople and ex-cannibals amidst the jaw-dropping biodiversity of New Guinea’s rainforest. Here they captured two new bird species, each of which has developed the ability to consume toxic food and turn that into a poison of their own.
The other new poisonous bird discovered is the The regent whistler (Pachycephala schlegelii). The two birds that the researchers discovered to be poisonous are the regent whistler (Pachycephala schlegelii), a species that belongs to a family of birds with a wide distribution and easily recognizable song well-known from across the Indo-Pacific region, and the rufous-naped bellbird (Aleadryas rufinucha).
"We were really surprised to find these birds to be poisonous as no new poisonous bird species has been discovered in over two decades. Particularly, because these two bird species are so common in this part of the world," says Knud Jønsson.
Neurotoxin causes muscle spasms
Most people are familiar with South and Central America’s iconic poison dart frogs – especially the golden poison frog. These small, brightly colored amphibians can kill a human at the slightest touch. The discovery of the two new poisonous bird species in New Guinea, which carry the same type of toxin in their skin and feathers, demonstrates that the frog toxin is more widespread than once believed.
The name Batrachotoxin comes from the Greek word for frog – Batrachos – and therefore means frog toxin. It earned its name after being discovered in poison dart frogs. It is one of the most powerful known neurotoxins and 250 times more toxic than strychnine.
Batrachotoxin works by forcing sodium channels in skeletal muscles to lock in an open position. This causes muscles to go into extreme spasm and can lead to death.
The new UCPH research demonstrates that animals which adapt to the toxin probably require changes in the voltage-gated sodium (NaV) channels where the toxin binds. The birds have mutations in the same areas as golden poison frog, but the specific changes are different.
The poison in these birds’ bodies and plumage is called Batrachotoxin. It is an incredibly potent neurotoxin that, in higher concentrations, such as those found in the skin of golden poison frogs, leads to muscle cramps and cardiac arrest nearly immediately after contact.
"The bird’s toxin is the same type as that found in frogs, which is a neurotoxin that, by forcing sodium channels in skeletal muscle tissue to remain open, can cause violent convulsions and ultimately death," explains Kasun Bodawatta.

|
Scooped by
Dr. Stefan Gruenwald
August 2, 2023 5:02 PM
|
Researchers have discovered new types of cells in the brain of developing flies thanks to a new method that creates cell-type-specific genetic tools. The study, published in the journal Proceedings of the National Academy of Sciences (PNAS), combines single-cell sequencing data with a novel algorithm to identify pairs of genes that point to previously unknown cells in the brains of fruit flies. Fruit flies (also known as Drosophila) have long been used as a model organism to study fundamental questions about the development and function of the brain. Instead of the 86 billion neurons found in humans, fruit flies have about 100,000 neurons -- making research into the brain a more manageable, yet still complex, endeavor. The use of genetic tools that can distinguish different types of cells in fruit flies has revolutionized the study of neural circuits in the brain, allowing scientists to understand circuit development, function, and behavior in a precise manner. "A hallmark of the central nervous system is the diversity of different cell types that are responsible for so many different functions," said Claude Desplan, Silver Professor of Biology and Neural Science at NYU and the study's senior author. Previous research in Desplan's lab used single-cell sequencing to determine that there are approximately 200 cell types in the developing fly's visual system. Single-cell sequencing reveals gene expression, so when cells have the same gene expression patterns, they are likely doing the same job and are therefore the same cell type. Scientists were able to identify roughly half of the 200 cell types in the developing fly's visual system based on their gene expression and prior studies, but they lacked a way to more easily study and label the other 100 cell types. Existing tools that allowed precise manipulation of neural circuits of adult fruit flies often failed to label the same neurons during development, rendering these tools unfit to study cells in the developing brain. "Moreover, the previous approach to identifying cell types involves laborious testing of numerous gene candidate combinations. We knew we needed a much more efficient approach to label specific cell types, and were able to tap into the growing amount of single-cell sequencing data that is available," said Yu-Chieh David Chen, a postdoctoral associate in NYU's Department of Biology and the study's first author.

|
Scooped by
Dr. Stefan Gruenwald
July 28, 2023 3:11 PM
|
For the first time, scientists have managed to induce virgin birth in an animal that usually reproduces sexually: the fruit fly Drosophila melanogaster. Once induced in this fruit fly, this ability is passed on through the generations: the offspring can reproduce either sexually if there are males around, or by virgin birth if there aren't. For most animals, reproduction is sexual -- it involves a female's egg being fertilized by a male's sperm. Virgin birth, or 'parthenogenesis', is the process by which an egg develops into an embryo without fertilization by sperm -- a male is not needed. The offspring of a virgin birth are not exact clones of their mother but are genetically very similar, and are always female. "We are the first to show that you can engineer virgin births to happen in an animal -- it was very exciting to see a virgin fly produce an embryo able to develop to adulthood, and then repeat the process," said Dr Alexis Sperling, a researcher at the University of Cambridge and first author of the paper. She added: "In our genetically manipulated flies, the females waited to find a male for half their lives -- about 40 days -- but then gave up and proceeded to have a virgin birth." In the experiments, only 1-2% of the second generation of female flies with the ability for virgin birth produced offspring, and this occurred only when there were no male flies around. When males were available, the females mated and reproduced in the normal way. Switching to a virgin birth can be a survival strategy: a one-off generation of virgin births can help to keep the species going. The study has been published in the journal Current Biology. To achieve their results, researchers first sequenced the genomes of two strains of another species of fruit fly, called Drosophila mercatorum. One strain needs males to reproduce, the other reproduces only through virgin birth. They identified the genes that were switched on, or switched off, when the flies were reproducing without fathers. With the candidate genes for virgin birth ability identified in Drosophila mercatorum, the researchers altered what they thought were the corresponding genes in the model fruit fly, Drosophila melanogaster. It worked: The flies suddenly acquired the ability for virgin birth. The research involved over 220,000 virgin fruit flies and took six years to complete.Key to the discovery was the fact that this work was done in Drosophila melanogaster -- the researchers say it would have been incredibly difficult in any other animal. This fly has been the 'model organism' for research in genetics for over 100 years and its genes are very well understood. Sperling, who carried out this work in the Department of Genetics, has recently moved to Cambridge Crop Science Centre to work on crop pests and hopes to eventually investigate why virgin birth in insects may be becoming more common, particularly in pest species. "If there's continued selection pressure for virgin births in insect pests, which there seems to be, it will eventually lead to them reproducing only in this way. It could become a real problem for agriculture because females produce only females, so their ability to spread doubles," said Sperling. The females of some egg-laying animals -- including birds, lizards and snakes, can switch naturally to give birth without males. But virgin birth in animals that normally sexually reproduce is rare, often only observed in zoo animals, and usually happens when the female has been isolated for a long time and has little hope of finding a mate.

|
Scooped by
Dr. Stefan Gruenwald
July 10, 2023 3:09 PM
|
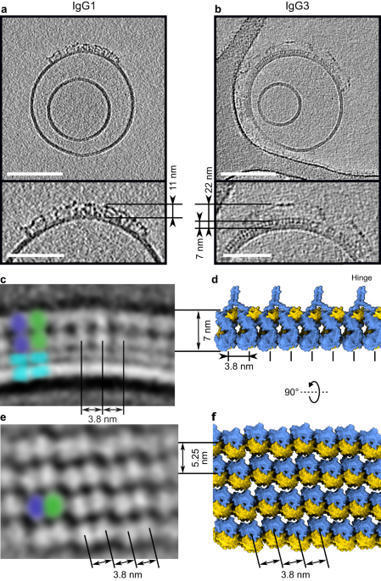
IgG3 antibodies are the most potent subclass of IgG, yet are the only subclass not used as therapeutics. That stems partly from the fact that nobody knew what they looked like and what complexes they formed and when. Now, Leoni Abendstein solved the structure of antigen-bound IgG3, both alone and activating complement, and discovered loads about how these unique antibodies work IgG3 is unique among the IgG subclasses due to its extended hinge, allotypic diversity and enhanced effector functions, including highly efficient pathogen neutralisation and complement activation. It is also underrepresented as an immunotherapeutic candidate, partly due to a lack of structural information. Here, we use cryoEM to solve structures of antigen-bound IgG3 alone and in complex with complement components. These structures reveal a propensity for IgG3-Fab clustering, which is possible due to the IgG3-specific flexible upper hinge region and may maximise pathogen neutralisation by forming high-density antibody arrays. IgG3 forms elevated hexameric Fc platforms that extend above the protein corona to maximise binding to receptors and the complement C1 complex, which here adopts a unique protease conformation that may precede C1 activation. Mass spectrometry reveals that C1 deposits C4b directly onto specific IgG3 residues proximal to the Fab domains. Structural analysis shows this to be caused by the height of the C1-IgG3 complex. Together, these data provide structural insights into the role of the unique IgG3 extended hinge, which will aid the development and design of upcoming immunotherapeutics based on IgG3.

|
Scooped by
Dr. Stefan Gruenwald
May 27, 2023 11:21 AM
|
For more than a century, biologists have wondered what the earliest animals were like when they first arose in the ancient oceans over half a billion years ago. Searching among today’s most primitive-looking animals for the earliest branch of the animal tree of life, scientists gradually narrowed the possibilities down to two groups: sponges, which spend their entire adult lives in one spot, filtering food from seawater; and comb jellies, voracious predators that oar their way through the world’s oceans in search of food. In a new study published this week in the journal Nature, researchers use a novel approach based on chromosome structure to come up with a definitive answer: Comb jellies, or ctenophores (teen’-a-fores), were the first lineage to branch off from the animal tree. Sponges were next, followed by the diversification of all other animals, including the lineage leading to humans. Although the researchers determined that the ctenophore lineage branched off before sponges, both groups of animals have continued to evolve from their common ancestor. Nevertheless, evolutionary biologists believe that these groups still share characteristics with the earliest animals, and that studying these early branches of the animal tree of life can shed light on how animals arose and evolved to the diversity of species we see around us today. “The most recent common ancestor of all animals probably lived 600 or 700 million years ago. It’s hard to know what they were like because they were soft-bodied animals and didn’t leave a direct fossil record. But we can use comparisons across living animals to learn about our common ancestors,” said Daniel Rokhsar, University of California, Berkeley professor of molecular and cell biology and co-corresponding author of the paper along with Darrin Schultz and Oleg Simakov of the University of Vienna. “It’s exciting — we’re looking back deep in time where we have no hope of getting fossils, but by comparing genomes, we’re learning things about these very early ancestors.” Understanding the relationships among animal lineages will help scientists understand how key features of animal biology, such as the nervous system, muscles and digestive tract, evolved over time, the researchers say. “We developed a new way to take one of the deepest glimpses possible into the origins of animal life,” said Schultz, the lead author and a former UC Santa Cruz graduate student and researcher at the Monterey Bay Aquarium Research Institute (MBARI) who is now a postdoctoral researcher at the University of Vienna. “This finding will lay the foundation for the scientific community to begin to develop a better understanding of how animals have evolved.” What is an animal?
Most familiar animals, including worms, flies, mollusks, sea stars and vertebrates — and including humans — have a head with a centralized brain, a gut running from mouth to anus, muscles and other shared features that had already evolved by the time of the famed “Cambrian Explosion” around 500 million years ago. Together, these animals are called bilaterians. Other bona fide animals, however, such as jellyfish, sea anemones, sponges and ctenophores, have simpler body plans. These creatures lack many bilaterian features — for example, they lack a defined brain and may not even have a nervous system or muscles — but still share the hallmarks of animal life, notably the development of multicellular bodies from a fertilized egg. The evolutionary relationships among these diverse creatures — specifically, the order in which each of the lineages branched off from the main trunk of the animal tree of life — has been controversial. With the rise of DNA sequencing, biologists were able to compare the sequences of genes shared by animals to construct a family tree that illustrates how animals and their genes evolved over time since the earliest animals arose in the Precambrian Period. But these phylogenetic methods based on gene sequences failed to resolve the controversy over whether sponges or comb jellies were the earliest branch of the animal tree, in part because of the deep antiquity of their divergence, Rokhsar said. “The results of sophisticated sequence-based studies were basically split,” he said. “Some researchers did well-designed analyses and found that sponges branched first. Others did equally complex and justifiable studies and got ctenophores. There hasn’t really been any convergence to a definitive answer.”

|
Scooped by
Dr. Stefan Gruenwald
May 23, 2023 11:00 PM
|
Scales, spines, feathers and hair are examples of vertebrate skin appendages, which constitute a remarkably diverse group of micro-organs. Despite their natural multitude of forms, these appendages share early developmental processes at the embryonic stage. Two researchers from the University of Geneva (UNIGE) have discovered how to permanently transform the scales that normally cover the feet of chickens into feathers, by specifically modifying the expression of certain genes. These results, published in the journal Science Advances, open new perspectives for studying mechanisms that have enabled radical evolutionary transitions in form among species.
The skin of terrestrial vertebrates is adorned with diverse keratinized appendages, such as hair, feathers, and scales. Despite the diversity of forms within and among species, the embryonic development of skin appendages typically begins in a very similar way. Indeed, all of these structures develop from cells that produce a localized thickening on the skin surface and express particular genes. One of these genes, called Sonic hedgehog (Shh), controls a signaling pathway - a communication system that allows the transmission of messages within and between cells. Shh signalling is involved in the development of diverse structures, including the neural tube, limb buds and skin appendages.
A common ancestor
The laboratory of Michel Milinkovitch, professor in the Department of Genetics and Evolution at the Faculty of Science of the UNIGE, is interested in the physical and biological processes that generate the diversity of skin appendages in vertebrates. In particular, his group has previously demonstrated that hair, feathers and scales are homologous structures inherited from a reptilian common ancestor.
Feathers of the chicken embryo are used by scientists as a model system to understand skin appendage development. While it is known that certain breeds of chickens, such as the ‘Brahma’ and ‘Sablepoot’ varieties, exhibit feathered legs and dorsal foot surfaces, the genetic determinism of this trait is not fully understood.
A transient modification for a permanent change
As the signaling pathways responsible for this transformation have not been fully determined, Michel Milinkovitch’s group investigated the potential role of the Shh pathway. “We used the classic technique of ‘egg candling’, in which a powerful torch illuminates blood vessels on the inside of the eggshell. This allowed us to precisely treat chicken embryos with a molecule that specifically activates the Shh pathway, injected directly into the bloodstream,’’ explains Rory Cooper, a post-doctoral researcher in Michel Milinkovitch’s laboratory and co-author of the study.
The two scientists observed that this single stage-specific treatment is sufficient to trigger the formation of abundant juvenile down-type feathers, in areas that would normally be covered with scales. Remarkably, these experimentally-induced feathers are comparable to those covering the rest of the body, as they are regenerative and are subsequently and autonomously replaced by adult feathers.
After comparison with embryos injected with a ‘control’ solution (without the active molecule), RNA sequencing analysis showed that the Shh pathway is both immediately and persistently activated following injection of the molecule. This confirms that activation of the Shh pathway underlies the conversion of scales into feathers.
‘‘Our results indicate that an evolutionary leap - from scales to feathers - does not require large changes in genome composition or expression. Instead, a transient change in expression of one gene, Shh, can produce a cascade of developmental events leading to the formation of feathers instead of scales,’’ says Michel Milinkovitch. This research, initially focused on the study of the development of scales and feathers, therefore has important implications for understanding the evolutionary mechanisms generating the enormous diversity of animal forms observed in nature.

|
Scooped by
Dr. Stefan Gruenwald
May 15, 2023 6:25 PM
|
Electron cryo-tomography (cryo-ET) is emerging as a powerful technique to provide detailed 3D images of cellular environments and enclosed biomolecules. However, one of the challenges of the methodology is the identification of protein molecules in the images for further processing. A research team around Stefan Raunser, Director at the MPI of Molecular Physiology in Dortmund, led by Thorsten Wagner, developed software to pick proteins in crowded cellular volumes. The new open-source tool, called TomoTwin, is based on deep metric learning and allows scientists to locate several proteins with high accuracy and throughput without manually creating or retraining the network each time. The paper is published in the journal Nature Methods. "TomoTwin paves the way for automated identification and localization of proteins directly in their cellular environment, expanding the potential of cryo-ET," says Gavin Rice, co-first author of the publication. Cryo-ET has the potential to decipher how biomolecules work within a cell and, by that, to unveil the basis of life and the origin of diseases. In a cryo-ET experiment, scientists use a transmission electron microscope to obtain 3D images, called tomograms, of the cellular volume containing complex biomolecules. To gain a more detailed image of each different protein, they average as many copies of them as possible—similar to photographers capturing the same photo at varying exposures to later combine them in a perfectly exposed image. Crucially, one has to correctly identify and locate the different proteins in the picture before averaging them. "Scientists can attain hundreds of tomograms per day, but we lacked tools to fully identify the molecules within them," says Rice. So far, researchers have used algorithms based on templates of already known molecular structures to search for matches in the tomograms, but these tend to be error-prone. Identifying molecules by hand is another option which ensures high-quality picking but takes days to weeks per dataset. Another possibility would be to use a form of supervised machine learning. These tools can be very accurate but currently lack usability, as they require manually labeling thousands of examples to train the software for each new protein, an almost impossible task for small biological molecules in a crowded cellular environment. TomoTwin The newly developed software TomoTwin overcomes many of these obstacles: It learns to pick the molecules that are similar in shape within a tomogram and maps them to a geometric space—the system is rewarded for placing similar proteins near each other and penalized otherwise. In the new map researchers can isolate and accurately identify the different proteins and use this to locate them inside the cell. "One advantage of TomoTwin is that we provide a pre-trained picking model," says Rice. By removing the training step, the software can even run on local computers—where processing a tomogram usually requires 60-90 minutes, runtime on the MPI supercomputer Raven is reduced to 15 minutes per tomogram. TomoTwin allows researchers to pick dozens of tomograms in the time it takes to manually pick a single one, therefore increasing the throughput of data and the averaging rate to obtain a better image. The software can currently locate globular proteins or protein complexes larger than 150 kilodaltons in cells; in the future, the Raunser group aims to include membrane proteins, filamentous proteins, and proteins of smaller sizes.

|
Rescooped by
Dr. Stefan Gruenwald
from Virus World
May 1, 2023 12:08 PM
|
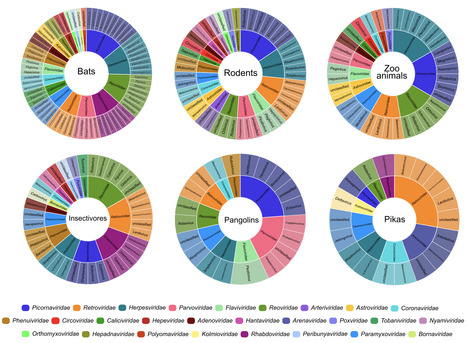
Wildlife is reservoir of emerging viruses. Here we identified 27 families of mammalian viruses from 1981 wild animals and 194 zoo animals collected from south China between 2015 and 2022, isolated and characterized the pathogenicity of eight viruses. Bats harbor high diversity of coronaviruses, picornaviruses and astroviruses, and a potentially novel genus of Bornaviridae. In addition to the reported SARSr-CoV-2 and HKU4-CoV-like viruses, picornavirus and respiroviruses also likely circulate between bats and pangolins. Pikas harbor a new clade of Embecovirus and a new genus of arenaviruses. Further, the potential cross-species transmission of RNA viruses (paramyxovirus and astrovirus) and DNA viruses (pseudorabies virus, porcine circovirus 2, porcine circovirus 3 and parvovirus) between wildlife and domestic animals was identified, complicating wildlife protection and the prevention and control of these diseases in domestic animals. This study provides a nuanced view of the frequency of host-jumping events, as well as assessments of zoonotic risk. Monitoring the diversity of viruses infecting animals is important for assessing zoonotic risk. Here, the authors use metatranscriptomics to characterise the viromes of small mammals, pangolins, and zoo animals in China to identify potentially zoonotic viruses. Published in Nature Comm. (April 29, 2023): https://doi.org/10.1038/s41467-023-38202-4
Via Juan Lama
With chatbots like ChatGPT making a splash, machine learning is playing an increasingly prominent role in our lives. For many of us, it’s been a mixed bag. We rejoice when our Spotify For You playlist finds us a new jam, but groan as we scroll through a slew of targeted ads on our Instagram feeds. Machine learning is also changing many fields that may seem surprising. One example is my discipline, ornithology, the study of birds. It isn’t just solving some of the biggest challenges associated with studying bird migration; more broadly, machine learning is expanding the ways in which people engage with birds. As spring migration picks up, here’s a look at how machine learning is influencing ways to research birds and, ultimately, to protect them. The challenge of conserving migratory birds Most birds in the Western Hemisphere migrate twice a year, flying over entire continents between their breeding and nonbreeding grounds. While these journeys are awe-inspiring, they expose birds to many hazards en route. These include extreme weather, food shortages and light pollution that can attract birds and cause them to collide with buildings. Our ability to protect migratory birds is only as good as the science that tells us where they go. And that science has come a long way. Banding to track bird migration In 1920, the U.S. Geological Survey launched the Bird Banding Laboratory, spearheading an effort to put bands with unique markers on birds, then recapture the birds in new places to figure out where they traveled. Today researchers can deploy a variety of lightweight tracking tags on birds to discover their migration routes. These tools have uncovered the spatial patterns of where and when birds of many species migrate. However, tracking birds has limitations. For one thing, over 4 billion birds migrate across the continent every year. Even with increasingly affordable equipment, the number of birds that we track is a drop in the bucket. And even within a species, migratory behavior may vary across sexes or populations. Further, tracking data tells us where birds have been, but it doesn’t necessarily tell us where they’re going. Migration is dynamic, and the climates and landscapes that birds fly through are constantly changing. That means it’s crucial to be able to predict their movements. Using machine learning to forecast bird migration This is where machine learning comes in. Machine learning is a subfield of artificial intelligence that gives computers the ability to learn tasks or associations without explicitly being programmed. We use it to train algorithms that tackle various tasks, from forecasting weather to predicting March Madness upsets. But applying machine learning requires data. And the more data the better. Luckily, scientists have inadvertently compiled decades of data on migrating birds through the Next Generation Weather Radar system. This network, known as NEXRAD, measures weather dynamics and helps predict future weather events. But it also picks up signals from birds as they fly through the atmosphere.
Via LIGHTING

|
Scooped by
Dr. Stefan Gruenwald
April 4, 2023 7:56 PM
|
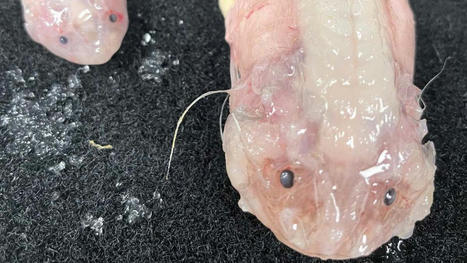
Scientists have set a new record by filming a deep sea snailfish (Pseudoliparis belyaevi) at 8,000 meters below the surface of the ocean. These snailfish specimens are the deepest fish ever caught, recovered from a depth of 8022m in the Japan Trench. Scientists have delved eight kilometers below the ocean’s surface to film a fish, and set new records in the process. While exploring the Japan, Izu-Ogasawara and Ryukyu trenches as part of a decade-long study into the world’s deep sea fish populations, teams from the Universities of Western Australia and Tokyo University of Marine Science and Technology spotted a snailfish (genus Pseudoliparis) cruising at a depth of 8,366 meters. It’s the deepest recording of a fish yet taken by humans. Pseudoliparis are a genus first described midway through last century, and known to inhabit the farthest depths of the Pacific Ocean’s deep trenches. They are ray-finned fishes and part of the same animal order as lionfishes. The records weren’t over, however, with the study team trapping two Pseudoliparis belyaevi at 8,002 meters – a record catch depth for this species. “It is not necessarily that they are living at 8,336m but rather we have enough information on this environment to have predicted that these trenches would be where the deepest fish would be,” says the study’s chief scientist, Professor Alan Jamieson.“ Until this expedition, no one had ever seen nor collected a single fish from this entire trench. “We have spent over 15 years researching these deep snailfish; there is so much more to them than simply the depth, but the maximum depth they can survive is truly astonishing. In other trenches such as the Mariana Trench, we were finding them at increasingly deeper depths just creeping over that 8,000m mark in fewer and fewer numbers, but around Japan they are really quite abundant.”

|
Scooped by
Dr. Stefan Gruenwald
March 25, 2023 12:37 PM
|
Prof. Dr. Stephen Elledge's lab has applied genetic tools to uncover how viruses exploit mammalian proteins to further their goals of reproduction and how the cell combats these viruses. They have identified a large number of host factors required for a wide variety of viruses to replicate and factors that restrain this replication. With this work, they hope that by understanding these pathways, they can inform antiviral drug discovery and host-pathogen biology. They have also initiated a chemical screening effort to explore how existing FDA approved drugs can be used in combination to control acute viral infections.

|
Scooped by
Dr. Stefan Gruenwald
March 6, 2023 8:58 PM
|
Aber-clam Lincoln, a quahog clam believed to be 214 years old found at Alligator Point, was released into the Gulf of Mexico Friday by his caretakers at the Gulf Specimen Marine Lab in Panacea. Americorps member Blaine Parker dug up the two-century old mollusk while collecting shellfish to make chowder. Parker said it is hefty enough to make two servings and has shells large enough to use as bowls to serve it in. “We were just going to eat it, but we thought about it a while and figured it was probably pretty special. So, we didn’t want to kill it,” said Parker. Instead, he took it to the aquarium at the Gulf Specimen Marine Lab where he works as a specimen collector. While most Ocean quahogs are within 2.8 to 4.3 inches in length and weigh up to a half pound, Lincoln checks in at 6 inches and weighs 2.6 pounds. The clam has been drawing up to 100 visitors a day. Parker is a recent Eckerd College graduate with a degree in environmental studies and marine science. He said, a clam, like a tree, lays down annual growth bands, alternating bands of light on its shell, that scientists used to estimate its age. There are 214 layers on the shell of the clam he found, which would mean it was likely born in 1809, the same year as Abraham Lincoln. It was also discovered on Presidents Day weekend. So, Parker named his find Aber-clam Lincoln. Quahogs occur in the Atlantic from Greenland south to North Carolina. They move about by being tossed in the surf, and crawl along the sand by expanding and contracting their muscle. With Lincoln being so old it is possible, like many Floridians, circumstances moved him south for him to become one of the oldest known Florida transplants. Quahogs are known to live 200 or more years. A 507-year-old clam, found in the Icelandic seabed and named Ming, broke the Guinness World Record for oldest animal when it was discovered in 2007. Scientists think an incredible low metabolic rates of Lincoln and other quahogs is responsible for their long life. A University of Kansas study of the energy needs of 299 species of extinct and living bivalves found those with minimal energy requirements escaped extinction.

|
Scooped by
Dr. Stefan Gruenwald
February 23, 2023 12:23 PM
|
Many migratory species use the Earth’s magnetic field to keep their journeys on track. Now a study of a very non-migratory animal, the Drosophila fruit fly, shows the same capacity exists in some unexpected places. Perhaps humans are the rare ones because we don’t have this capability; if so, why? In the quest for survival, access to information about the world, particularly information your rivals lack, is exceptionally valuable. So it is not surprising animals have developed an astonishing array of ways to observe the world around them. Magnetic fields are one of these, but before humanity’s invention of powerful electromagnets these were generally very weak. The effort required to detect them was much greater than for light or sound. Consequently, biologists thought that only those animals that really needed to know their place on Earth – migratory pigeons or turtles for example – had exploited magnetoreception. However, a paper in Nature calls this into question. The possibility that Drosophila are capable of magnetoreception was raised in 2015 with the identification of a MagR protein produced by the flies that orientates itself to align with magnetic fields. The newly-published paper goes past this, revealing two methods by which the flies’ cells appear able to detect fields. The previous work identified photoreceptor proteins known as cryptochromes as being the sensors used by Drosophila to detect fields, with the capacity apparently failing in flies engineered not to produce cryptochromes leaving them magnetically blind. The authors of the new paper point to work showing cryptochromes do this by harnessing the powers of quantum super-positioning. However, the team also question the need for cryptochromes, showing their role may be substituted by a molecule that occurs in all living cells, humans included. Dr Alex Jones (no, not that one) of the National Physics Laboratory said in a statement, "The absorption of light by the cryptochrome results in movement of an electron within the protein which, due to quantum physics, can generate an active form of cryptochrome that occupies one of two states. The presence of a magnetic field impacts the relative populations of the two states, which in turn influences the 'active-lifetime' of this protein." The authors showed the molecule flavin adenine dinucleotide (FAD) binds to the cryptochromes to create their sensitivity to magnetism. However, they also found the cryptochromes may be an amplifier of FAD’s capacity, not essential to it. Even without cryptochromes, fly cells engineered to express extra FAD were able to respond to the presence of magnetic fields, as well as being highly sensitive to blue light in the presence of these fields. The magnetoreception required nothing more complex than an electron transfer to a side chain. The authors think cryptochromes may have evolved to take advantage of this. "This study may ultimately allow us to better appreciate the effects that magnetic field exposure might potentially have on humans,” said co-lead author Professor Ezio Rosato of the University of Leicester.
|

|
Scooped by
Dr. Stefan Gruenwald
August 12, 2023 2:38 PM
|
Two recent papers have demonstrated that during a critical early period of brain development, the gut’s microbiome — the assortment of bacteria that grow within in it — helps to mold a brain system that’s important for social skills later in life. Scientists found this influence in fish, but molecular and neurological evidence plausibly suggests that some form of it could also occur in mammals, including humans. In a recent paper published in PLOS Biology, researchers found that zebra fish who grew up lacking a gut microbiome were far less social than their peers with colonized colons, and the structure of their brains reflected the difference. In a related article in BMC Genomics in late September, they described molecular characteristics of the neurons affected by the gut bacteria. Equivalents of those neurons appear in rodents, and scientists can now look for them in other species, including humans.

|
Rescooped by
Dr. Stefan Gruenwald
from Virus World
July 28, 2023 3:37 PM
|
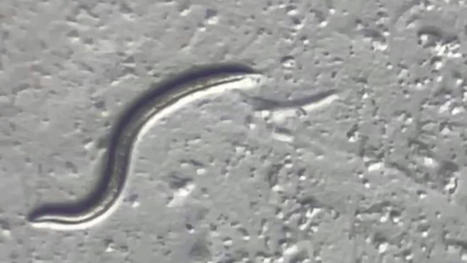
The animal is a previously unknown species that may help researchers unlock secrets of surviving harsh environments. A female microscopic roundworm that spent the last 46,000 years in suspended animation deep in the Siberian permafrost was revived and started having babies in a laboratory dish. By sequencing the genome of this Rip Van Winkle roundworm, scientists revealed it to be a new species of nematode, which is described in a study published Thursday in the journal PLOS Genetics. Nematodes today are among the most ubiquitous organisms on Earth, inhabiting the soil, the water and the ocean floor. “The vast majority of nematode species have not been described,” William Crow, a nematologist at the University of Florida who was not involved in the study, wrote in an email. The ancient Siberian worm could be a species that has since gone extinct, he said. “However, it very well could be a commonly occurring nematode that no one got around to describing yet." Published in Plos Genetics (July 27, 2023): https://doi.org/10.1371/journal.pgen.1010798
Via Juan Lama

|
Scooped by
Dr. Stefan Gruenwald
July 15, 2023 6:31 PM
|
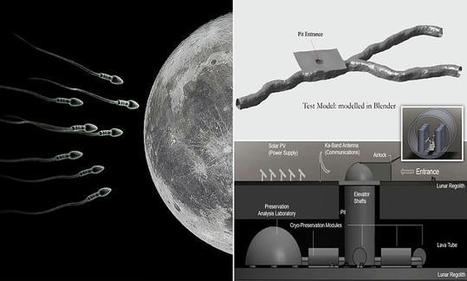
The lunar gene bank - which could also house seed and spore samples - is envisaged as being built under the lunar surface, in a hollow, cooled lava tube. Extinction insurance for planet earth is envisioned by sending sperm and egg samples from 6.7 million species to the moon for safekeeping. How could earth species be destroyed to require a “backup”? - To protect Earth’s species diversity, we presently store genetic material in vaults
- Material including seeds and spores could be used to recover lost species
- But gene banks on Earth are vulnerable to the catastrophes they insure against
- US experts propose to more safely store specimens in lava tubes on the moon
- The deposits would be cryogenically frozen and tended by levitating robots
The lunar gene bank — which could also house seed and spore samples — is envisaged as being built under the lunar surface, in a hollow, cooled lava tube. Specimens deposited in the ark would be kept refrigerated at cryogenic temperatures, with the facility powered by solar panels on the lunar surface. The ark would preserve Earth’s genetic diversity in the event of a global catastrophe, such as might be caused by climate change, a supervolcano or an asteroid impact. Earth is naturally a volatile environment,’ said study author and mechanical engineer Jekan Thanga of the University of Arizona. ‘As humans, we had a close call about 75,000 years ago with the Toba supervolcano eruption, which caused a 1,000-year cooling period and, according to some, aligns with an estimated drop in human diversity,’ he added. ‘Because human civilization has such a large footprint, if it were to collapse, that could have a negative cascading effect on the rest of the planet.’ The idea of creating gene banks to restore lost biodiversity in the future is not new — more than a million seed samples are currently stored in the Svalbard Global Seed Vault on the island of Spitsbergen in the Artic Sea, for example. However, Professor Thanga and colleagues explained that locating such facilities on Earth leaves them also vulnerable to accidental loss. Climate change, for example, has the potential to push many species into terminal decline in the future — and, at the same time, the rising sea levels which will accompany global warming will see the Svalbard vault lost beneath the waves. Removed but still accessible, the moon may present a safer location. Building a genetic storage facility on the moon would be a significant undertaking — but one that Professor Thanga says would be possible. Read full story here…

|
Rescooped by
Dr. Stefan Gruenwald
from Virus World
July 6, 2023 6:16 PM
|
Large DNA viruses of the phylum Nucleocytoviricota infect diverse eukaryotic hosts from protists to humans, with profound consequences for aquatic and terrestrial ecosystems. While nucleocytoviruses are known to be highly diverse in metagenomes, knowledge of their capsid structures is restricted to a few characterized representatives. Here, we visualize giant virus-like particles (VLPs, diameter >0.2 μm) directly from the environment using transmission electron microscopy. We found that Harvard Forest soils contain a higher diversity of giant VLP morphotypes than all hitherto isolated giant viruses combined. These included VLPs with icosahedral capsid symmetry, ovoid shapes similar to pandoraviruses, and bacilliform shapes that may represent novel viruses. We discovered giant icosahedral capsids with structural modifications that had not been described before including tubular appendages, modified vertices, tails, and capsids consisting of multiple layers or internal channels. Many giant VLPs were covered with fibers of varying lengths, thicknesses, densities, and terminal structures. These findings imply that giant viruses employ a much wider array of capsid structures and mechanisms to interact with their host cells than is currently known. We also found diverse tailed bacteriophages and filamentous VLPs, as well as ultra-small cells. Our study offers a first glimpse of the vast diversity of unexplored viral structures in soil and reinforces the potential of transmission electron microscopy for fundamental discoveries in environmental microbiology. Preprint available at bioRxiv (June 30, 2023): https://doi.org/10.1101/2023.06.30.546935
Via Juan Lama

|
Rescooped by
Dr. Stefan Gruenwald
from Virus World
May 26, 2023 11:59 AM
|
In a recent study posted to the bioRxiv* preprint server, researchers analyzed the viral genomes of 28 endemic viruses to study the evolution of the ability of viruses to evade the neutralizing antibodies elicited by vaccines or previous infections. Background Viruses evolve rapidly and adapt to changing environments due to their high mutation rates and low generation time. Often viruses adapted to different animal hosts infect humans and optimize the methods through which they enter and replicate in the host cell, increasing the human-to-human transmission and evolving into a novel pathogen. The early stages of pandemics are often characterized by high adaptive evolutionary rates, as was seen during the coronavirus disease 2019 (COVID-19) pandemic and outbreaks related to various other respiratory viruses. While some viruses become endemic after adapting to a new host and do not evolve further, other endemic viruses continue to adapt through antigenic evolution, resulting in an arms race between the virus and the human immune system. Since viruses that undergo antigenic evolution pose the risk of repeat infections and increase their ability to evade vaccine-induced immunity, understanding which viruses continue to undergo antigen evolution could help manage future disease outbreaks. About the study The present study used sequence data for each gene in 28 viral genomes to estimate adaptive evolutionary rates. These 28 viruses spanned ten families and were transmitted between humans through various modes. Viruses with potentially high antigenic evolution rates were identified based on the high evolutionary rates for the genes coding for receptor-binding proteins since the receptor-binding region is involved in antibody neutralization and harbors most mutations that allow antigenic escape. The adaptive evolutionary rates were calculated in terms of the number of adaptive mutations in each codon per year, which allowed the adaptive evolutionary rates to be compared across the various genes in the genome and across viruses. The adaptive evolutionary rates of three viruses that were known to undergo antigenic evolution — coronavirus 229E, influenza viruses A/H3N2, and influenza viruses B/Yam — were compared against the evolutionary rates of three antigenically stable viruses, hepatitis A, measles, and influenza C/Yamagata. To understand the patterns of adaptive evolution in recent years, the sequence data for 28 viruses that are pathogenic to humans were obtained and curated. These viruses included deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) viruses and were transmitted between humans through bodily fluids, blood, vectors, fecal-oral, and respiratory routes. The researchers only investigated endemic viruses since they were interested in understanding the antigenic evolution that occurs during the endemic phase and not the initial adaptive phase. The evolutionary rates of the receptor binding protein, which was expected to be highly variable across endemic viruses, and the polymerase gene, which was expected to be conserved, were compared across the 28 viruses. Since the evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been relatively recent, and the Omicron variant carried a large number of mutations indicating a single fixation event, SARS-CoV-2 was compared with ten other antigenically evolving viruses by comparing the amino-acid substitution rates in the receptor binding protein. Results The results reported that 10 of the 28 viruses undergo adaptive evolution resulting in the antigenic mutations that allow the viruses to escape the immunity induced by previous infections and vaccines. Furthermore, comparing amino-acid substitution rates between SARS-CoV-2 and other viruses revealed that SARS-CoV-2 is evolving and accumulating mutations that cause protein-coding changes at rates much higher than other endemic viruses. Antigenic evolution was found to be more prevalent in RNA viruses. Still, the researchers believe that since the list of viruses included in the study was not comprehensive and comprised only well-studied viruses, determining the proportion of antigenically evolving endemic viruses is difficult. Furthermore, the rate of adaptive mutations might not reflect the phenotypic changes occurring in the viruses, implying that viruses with receptor-binding protein genes that evolve at the same rates might not develop the ability to evade vaccine or infection-induced immunity at the same rates. Conclusions Overall, the findings suggested that many human viruses that have become endemic continue to evolve antigenically, gaining the ability to escape the neutralizing antibodies generated by vaccinations or previous infections. Ten of the 28 viruses investigated in this study showed continued adaptive evolution. In contrast, the amino-acid substitution rates showed that SARS-CoV-2 is evolving faster than the other ten endemic human viruses. Cited ressearch available in bioRxiv (May 22, 2023): https://doi.org/10.1101/2023.05.19.541367
Via Juan Lama

|
Scooped by
Dr. Stefan Gruenwald
May 16, 2023 2:16 PM
|
Imagine using your cellphone to control the activity of your own cells to treat injuries and disease. It sounds like something from the imagination of an overly optimistic science fiction writer. But this may one day be a possibility through the emerging field of quantum biology. Over the past few decades, scientists have made incredible progress in understanding and manipulating biological systems at increasingly small scales, from protein folding to genetic engineering. And yet, the extent to which quantum effects influence living systems remains barely understood. Quantum effects are phenomena that occur between atoms and molecules that can't be explained by classical physics. It has been known for more than a century that the rules of classical mechanics, like Newton's laws of motion, break down at atomic scales. Instead, tiny objects behave according to a different set of laws known as quantum mechanics. For humans, who can only perceive the macroscopic world, or what's visible to the naked eye, quantum mechanics can seem counterintuitive and somewhat magical. Things you might not expect happen in the quantum world, like electrons "tunneling" through tiny energy barriers and appearing on the other side unscathed, or being in two different places at the same time in a phenomenon called superposition. Research in quantum mechanics is usually geared toward technology. However, and somewhat surprisingly, there is increasing evidence that nature—an engineer with billions of years of practice—has learned how to use quantum mechanics to function optimally. If this is indeed true, it means that our understanding of biology is radically incomplete. It also means that we could possibly control physiological processes by using the quantum properties of biological matter. Quantumness in biology is probably real Researchers can manipulate quantum phenomena to build better technology. In fact, you already live in a quantum-powered world: from laser pointers to GPS, magnetic resonance imaging and the transistors in your computer—all these technologies rely on quantum effects. In general, quantum effects only manifest at very small length and mass scales, or when temperatures approach absolute zero. This is because quantum objects like atoms and molecules lose their "quantumness" when they uncontrollably interact with each other and their environment. In other words, a macroscopic collection of quantum objects is better described by the laws of classical mechanics. Everything that starts quantum dies classical. For example, an electron can be manipulated to be in two places at the same time, but it will end up in only one place after a short while—exactly what would be expected classically. In a complicated, noisy biological system, it is thus expected that most quantum effects will rapidly disappear, washed out in what the physicist Erwin Schrödinger called the "warm, wet environment of the cell." To most physicists, the fact that the living world operates at elevated temperatures and in complex environments implies that biology can be adequately and fully described by classical physics: no funky barrier crossing, no being in multiple locations simultaneously. Chemists, however, have for a long time begged to differ. Research on basic chemical reactions at room temperature unambiguously shows that processes occurring within biomolecules like proteins and genetic material are the result of quantum effects. Importantly, such nanoscopic, short-lived quantum effects are consistent with driving some macroscopic physiological processes that biologists have measured in living cells and organisms. Research suggests that quantum effects influence biological functions, including regulating enzyme activity, sensing magnetic fields, cell metabolism and electron transport in biomolecules.

|
Scooped by
Dr. Stefan Gruenwald
May 5, 2023 12:57 PM
|
Newly discovered electrical activity within cells could change the way researchers think about biological chemistry The human body relies heavily on electrical charges. Lightning-like pulses of energy fly through the brain and nerves and most biological processes depend on electrical ions traveling across the membranes of each cell in our body. These electrical signals are possible, in part, because of an imbalance in electrical charges that exists on either side of a cellular membrane. Until recently, researchers believed the membrane was an essential component to creating this imbalance. But that thought was turned on its head when researchers at Stanford University discovered that similar imbalanced electrical charges can exist between microdroplets of water and air. Now, researchers at Duke University have discovered that these types of electric fields also exist within and around another type of cellular structure called biological condensates. Like oil droplets floating in water, these structures exist because of differences in density. They form compartments inside the cell without needing the physical boundary of a membrane. Inspired by previous research demonstrating that microdroplets of water interacting with air or solid surfaces create tiny electrical imbalances, the researchers decided to see if the same was true for small biological condensates. They also wanted to see if these imbalances sparked reactive oxygen, “redox,” reactions like these other systems. Appearing on April 28, 2023 in the journal Chem, their foundational discovery could change the way researchers think about biological chemistry. It could also provide a clue as to how the first life on Earth harnessed the energy needed to arise. “In a prebiotic environment without enzymes to catalyze reactions, where would the energy come from?” asked Yifan Dai, a Duke postdoctoral researcher working in the laboratory of Ashutosh Chilkoti, the Alan L. Kaganov Distinguished Professor of Biomedical Engineering and Lingchong You, the James L. Meriam Distinguished Professor of Biomedical Engineering. “This discovery provides a plausible explanation of where the reaction energy could have come from, just as the potential energy that is imparted on a point charge placed in an electric field,” Dai said. When electric charges jump between one material and another, they can produce molecular fragments that can pair up and form hydroxyl radicals, which have the chemical formula OH. These can then pair again to form hydrogen peroxide (H2O2) in tiny but detectable amounts. “But interfaces have seldom been studied in biological regimes other than the cellular membrane, which is one of the most essential part of biology,” said Dai. “So we were wondering what might be happening at the interface of biological condensates, that is, if it is an asymmetric system too. These findings suggest why condensates are so important in the functioning of cells.” Cells can build biological condensates to either separate or trap together certain proteins and molecules, either hindering or promoting their activity. Researchers are just beginning to understand how condensates work and what they could be used for. Because the Chilkoti laboratory specializes in creating synthetic versions of naturally occurring biological condensates, the researchers were easily able to create a test bed for their theory. After combining the right formula of building blocks to create minuscule condensates, with help from postdoctoral scholar Marco Messina in? Christopher J. Chang’s group at the University of California – Berkeley, they added a dye to the system that glows in the presence of reactive oxygen species. Their hunch was right. When the environmental conditions were right, a solid glow started from the edges of the condensates, confirming that a previously unknown phenomenon was at work. Dai next talked with Richard Zare, the Marguerite Blake Wilbur Professor of Chemistry at Stanford, whose group established the electric behavior of water droplets. Zare was excited to hear about the new behavior in biological systems, and started to work with the group on the underlying mechanism. “Inspired by previous work on water droplets, my graduate student, Christian Chamberlayne, and I thought that the same physical principles might apply and promote redox chemistry, such as the formation of hydrogen peroxide molecules,” Zare said. “These findings suggest why condensates are so important in the functioning of cells.”

|
Scooped by
Dr. Stefan Gruenwald
April 24, 2023 12:42 PM
|
A new Griffith University study has found that humpback whales will use sandy, shallow bay areas to ‘roll’ around in sandy substrates to remove dead skin cells on their return journeys south to cooler waters. Marine ecologist Dr Olaf Meynecke, from the Griffith-led Whales and Climate Research Program and Coastal and Marine Research Centre, used suction cup tags to track southward migrating whales between August 2021 and October 2022. The CATS tags are fitted with integrated high-definition video, magnesium release system, a VHF transmitter for retrieval, magnetometers, a hydrophone and light, pressure, temperature and GPS sensors. Using data and footage collected from the tags, whales were observed performing full and side rolls in up to 49m water depth on the sea floor that was lined with fine sand or rubble. “On all occasions of sand rolling, the whales were observed on video to be slowly moving forward with their head first into the sand followed by rolling to one side or a full roll,” Dr Meynecke said. “During the different deployments, the sand rolling was observed in the context of socializing. The behavior was either following courtship, competition or other forms of socializing. So we believe that the whales exfoliate using the sand to assist with moulting and removal of ectoparasites such as barnacle and specifically select areas suitable for this behavior.” In tropical and subtropical waters, barnacles attach to the whales in their early life stages, and whales need to remove barnacles frequently to avoid excessive growth that leads to drag and energy loss. “Humpback whales host diverse communities of skin bacteria that can pose a threat for open wounds if bacteria grow in large numbers,” Dr Meynecke said. “Removing excess skin is likely a necessity to maintain a healthy bacterial skin community. Humpback whales can remove some barnacles and skin through surface activity such as breaches but not all.” Skin from the whales was observed to be falling off during the process of all identified rolls, and fish such as juvenile silver trevally were seen to be actively feeding from the whales’ skin during this behaviour similar to cleaning stations are coral reefs.

|
Scooped by
Dr. Stefan Gruenwald
April 7, 2023 10:55 AM
|
Multicellular organisms typically develop from a single cell into a collection of cells that all have the same genetic material. A research team now discovered a deviation from this developmental hallmark in the yellow crazy ant, Anoplolepis gracilipes. Males are chimeras of haploid cells from two divergent lineages: R and W. R cells are overrepresented in the males’ somatic tissues, whereas W cells are overrepresented in their sperm. Chimerism occurs when parental nuclei bypass syngamy and divide separately within the same egg. When syngamy takes place, the diploid offspring either develops into a queen when the oocyte is fertilized by an R sperm or into a worker when fertilized by a W sperm. This study reveals a mode of reproduction that may be associated with a conflict between lineages to preferentially enter the germ line.

|
Rescooped by
Dr. Stefan Gruenwald
from Virus World
March 30, 2023 3:33 PM
|
An Australian company resurrects flesh of lost species to demonstrate potential of meat grown from cells as food source. Recently, they created a mammoth meatball, resurrecting the flesh of the long-extinct animals. The project aims to demonstrate the potential of meat grown from cells, without the slaughter of animals, and to highlight the link between large-scale livestock production and the destruction of wildlife and the climate crisis. The mammoth meatball was produced by Vow, an Australian company, which is taking a different approach to cultured meat. There are scores of companies working on replacements for conventional meat, such as chicken, pork and beef. But Vow is aiming to mix and match cells from unconventional species to create new kinds of meat. The company has already investigated the potential of more than 50 species, including alpaca, buffalo, crocodile, kangaroo, peacocks and different types of fish. The first cultivated meat to be sold to diners will be Japanese quail, which the company expects will be in restaurants in Singapore this year. “We have a behaviour change problem when it comes to meat consumption,” said George Peppou, CEO of Vow . “The goal is to transition a few billion meat eaters away from eating [conventional] animal protein to eating things that can be produced in electrified systems. “And we believe the best way to do that is to invent meat. We look for cells that are easy to grow, really tasty and nutritious, and then mix and match those cells to create really tasty meat.” Tim Noakesmith, who cofounded Vow with Peppou, said: “We chose the woolly mammoth because it’s a symbol of diversity loss and a symbol of climate change.” The creature is thought to have been driven to extinction by hunting by humans and the warming of the world after the last ice age. The initial idea was from Bas Korsten at creative agency Wunderman Thompson: “Our aim is to start a conversation about how we eat, and what the future alternatives can look and taste like. Cultured meat is meat, but not as we know it.” Plant-based alternatives to meat are now common but cultured meat replicates the taste of conventional meat. Cultivated meat – chicken from Good Meat – is currently only sold to consumers in Singapore, but two companies have now passed an approval process in the US. In 2018, another company used DNA from an extinct animal to create gummy bears made from gelatine from a mastodon, another elephant-like animal. Vow worked with Prof Ernst Wolvetang, at the Australian Institute for Bioengineering at the University of Queensland, to create the mammoth muscle protein. His team took the DNA sequence for mammoth myoglobin, a key muscle protein in giving meat its flavor, and filled in the few gaps using elephant DNA. This sequence was placed in myoblast stem cells from a sheep, which replicated to grow to the 20bn cells subsequently used by the company to grow the mammoth meat. “It was ridiculously easy and fast,” said Wolvetang. “We did this in a couple of weeks. Initially, the idea was to produce dodo meat, he said, but the DNA sequences needed do notvyet exist!"
Via Juan Lama

|
Scooped by
Dr. Stefan Gruenwald
March 16, 2023 4:21 PM
|
Breaking a longstanding impasse in our understanding of olfaction, scientists at UC San Francisco (UCSF) have created the first molecular-level, 3D picture of how an odor molecule activates a human odorant receptor, a crucial step in deciphering the sense of smell. The findings, appearing online March 15, 2023, in the journal Nature, are poised to reignite interest in the science of smell with implications for fragrances, food science, and beyond. Odorant receptors -- proteins that bind odor molecules on the surface of olfactory cells -- make up half of the largest, most diverse family of receptors in our bodies; A deeper understanding of them paves the way for new insights about a range of biological processes. "This has been a huge goal in the field for some time," said Aashish Manglik, MD, PhD, an associate professor of pharmaceutical chemistry and a senior author of the study. The dream, he said, is to map the interactions of thousands of scent molecules with hundreds of odorant receptors, so that a chemist could design a molecule and predict what it would smell like. "But we haven't been able to make this map because, without a picture, we don't know how odor molecules react with their corresponding odor receptors," Manglik said. A Picture Paints the Scent of Cheese Smell involves about 400 unique receptors. Each of the hundreds of thousands of scents we can detect is made of a mixture of different odor molecules. Each type of molecule may be detected by an array of receptors, creating a puzzle for the brain to solve each time the nose catches a whiff of something new. "It's like hitting keys on a piano to produce a chord," said Hiroaki Matsunami, PhD, professor of molecular genetics and microbiology at Duke University and a close collaborator of Manglik. Matsunami's work over the past two decades has focused on decoding the sense of smell. "Seeing how an odorant receptor binds an odorant explains how this works at a fundamental level." To create that picture, Manglik's lab used a type of imaging called cryo-electron microscopy (cryo-EM), that allows researchers to see atomic structure and study the molecular shapes of proteins. But before Manglik's team could visualize the odorant receptor binding a scent molecule, they first needed to purify a sufficient quantity of the receptor protein. Odorant receptors are notoriously challenging, some say impossible, to make in the lab for such purposes. The Manglik and Matsunami teams looked for an odorant receptor that was abundant in both the body and the nose, thinking it might be easier to make artificially, and one that also could detect water-soluble odorants. They settled on a receptor called OR51E2, which is known to respond to propionate -- a molecule that contributes to the pungent smell of Swiss cheese. But even OR51E2 proved hard to make in the lab. Typical cryo-EM experiments require a milligram of protein to produce atomic-level images, but co-first author Christian Billesbøelle, PhD, a senior scientist in the Manglik Lab, developed approaches to use only 1/100th of a milligram of OR51E2, putting the snapshot of receptor and odorant within reach. "We made this happen by overcoming several technical impasses that have stifled the field for a long time," said Billesbøelle. "Doing that allowed us to catch the first glimpse of an odorant connecting with a human odorant receptor at the very moment a scent is detected." This molecular snapshot showed that propionate sticks tightly to OR51E2 thanks to a very specific fit between odorant and receptor. The finding jibes with one of the duties of the olfactory system as a sentinel for danger. While propionate contributes to the rich, nutty aroma of Swiss cheese, on its own, its scent is much less appetizing. "This receptor is laser focused on trying to sense propionate and may have evolved to help detect when food has gone bad," said Manglik. Receptors for pleasing smells like menthol or caraway might instead interact more loosely with odorants, he speculated.

|
Scooped by
Dr. Stefan Gruenwald
February 23, 2023 12:31 PM
|
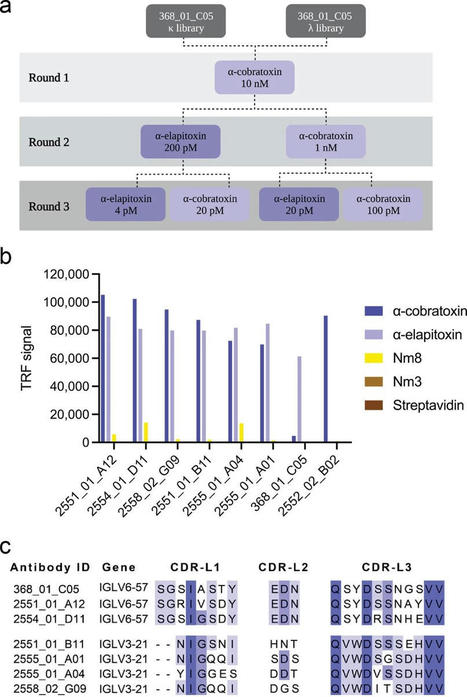
Snakes bite 5.4 million people each year—and roughly half are injected with venom, according to the WHO. Between 81,000 and 138,000 people die, while around three times as many suffer amputations and other permanent disabilities. Due to their size, children often suffer the most severe effects. For 128 years, our primary treatment against snakebite has been using mixtures of polyclonal antibodies derived from immunized animal blood. Although they are proven effective, these medicines may cause adverse reactions that can sometimes be severe. So, the search for novel ways to treat severe snakebite envenoming is ongoing. Recently, an international team of scientists led by DTU reached remarkable results and developed a new modernized prototype treatment that proves effective against the venom of African and Asian elapid snakes, such as some cobra, mamba, and krait species—many of which are among the world's deadliest. "We have previously developed antibodies against the venom toxins from single snake species; however, our new results demonstrate that our technology has great potential in neutralizing toxins from multiple species, even from different continents. This broadened cross-neutralization capacity is very promising. It could provide the basis for more effective treatments for snakebite victims in the future," says Andreas Hougaard Laustsen-Kiel, a professor at DTU Bioengineering. He conducted the research with colleagues at DTU, ETH Zurich, Universidad de Costa Rica, and industrial partners Sophion Bioscience and IONTAS. Their work is published in Nature Communications. New antibody works against several neurotoxins In essence, their approach is to develop antibodies of fully human origin, which offer fewer adverse reactions, competitive costs, and, when fine-tuned, superior efficacy. They use phage display technology, a popular in vitro methodology within drug discovery, to select antibodies that bind well to the toxins in the venom, enabling broad neutralization. "There has been a revolution in recombinant antibody technology over the last three decades. I am delighted to be involved in these efforts to direct phage display technology to the blight of snakebite envenomation," says John McCafferty, the inventor of antibody phage display. He founded IONTAS and has recently established a new anti-venom group at the University of Cambridge.
|




 Your new post is loading...
Your new post is loading...