Your new post is loading...
 Your new post is loading...
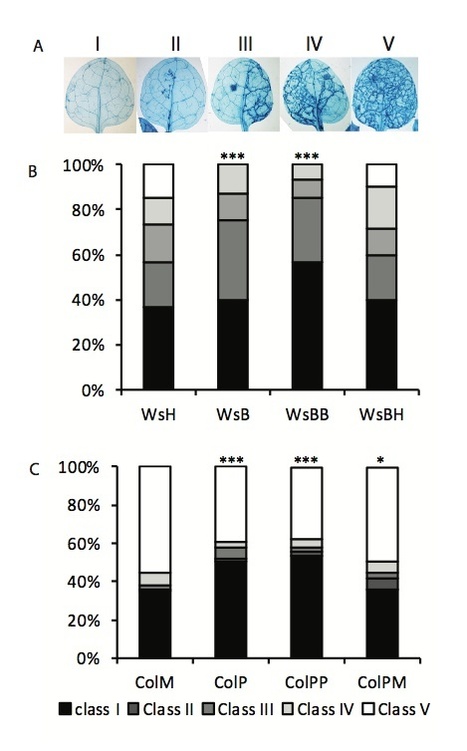
An attack of plants by pathogens or treatment with certain resistance-inducing compounds can lead to the establishment of a unique primed state of defense. Primed plants show enhanced defense reactions upon further challenge with biotic or abiotic stress. Here, we report that the primed state in Arabidopsis thaliana is still functional in the next generation without additional treatment. We compared the reactions of Arabidopsis plants that had been either primed with β-amino-butyric acid (BABA) or with an avirulent isolate of the bacteria Pseudomonas syringae pv tomato (PstavrRpt2). The descendants of primed plants showed a faster and higher accumulation of transcripts of defense-related genes in the SA-signaling pathway and enhanced disease resistance upon challenge inoculation with a virulent isolate of Pseudomonas syringae (Pst). In addition, the progeny of primed plants was also more resistant against the oomycete pathogen Hyaloperonospora arabidopsidis. When transgenerationally primed plants were subjected to an additional priming treatment, their descendants displayed an even stronger primed phenotype, suggesting that plants can inherit a sensitization for the priming phenomenon. Interestingly, this “primed to be primed” phenotype was much reduced in the Arabidopsis BABA-priming mutant ibs1. Our results demonstrate that the primed state of plants is transferred to their progeny and confers improved protection from pathogen attack as compared to the descendants of unprimed plants.
Secreted papain-like Cys proteases are important players in plant immunity. We previously reported that the C14 protease of tomato is targeted by cystatin-like EPIC proteins that are secreted by the oomycete pathogen Phytophthora infestans (Pinf) during infection. C14 has been under diversifying selection in wild potato species coevolving with Pinf and reduced C14 levels result in enhanced susceptibility for Pinf. Here, we investigated the role C14-EPIC-like interactions in the natural pathosystem of Arabidopsis with the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa). In contrast to the Pinf-solanaceae pathosystem, the C14 orthologous protease of Arabidopsis, RD21, does not evolve under diversifying selection in Arabidopsis, and rd21 null mutants do not show phenotypes upon compatible and incompatible Hpa interactions, despite the evident lack of a major leaf protease. Hpa isolates express highly conserved EPIC-like proteins during infections, but it is unknown if these HpaEPICs can inhibit RD21 and one of these HpaEPICs even lacks the canonical cystatin motifs. The rd21 mutants are unaffected in compatible and incompatible interactions with Pseudomonas syringae pv. tomato, but are significantly more susceptible for the necrotrophic fungal pathogen Botrytis cinerea, demonstrating that RD21 provides immunity to a necrotrophic pathogen.
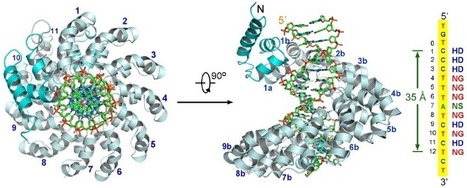
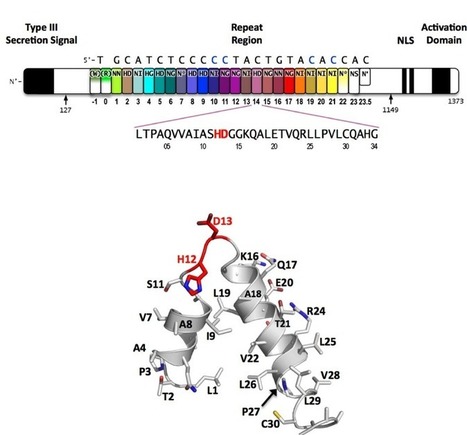
TAL effectors, secreted by phytopathogenic bacteria, recognize host DNA sequences through a central domain of tandem repeats. Each repeat comprises 33 to 35 conserved amino acids and targets a specific base pair using two hypervariable residues (known as RVD) at positions 12 and 13. Here, we report the crystal structures of a 11.5-repeat TAL effector in both DNA-free and DNA-bound states. Each TAL repeat comprises two helices connected by a short RVD-containing loop. The 11.5 repeats form a right-handed, super-helical structure that tracks along the sense strand of DNA duplex, with RVDs contacting the major groove. The 12th residue stabilizes the RVD loop, whereas the 13th residue makes a base-specific contact. Understanding DNA recognition by TAL effectors may facilitate rational design of DNA-binding proteins with biotechnological applications.
Via dromius
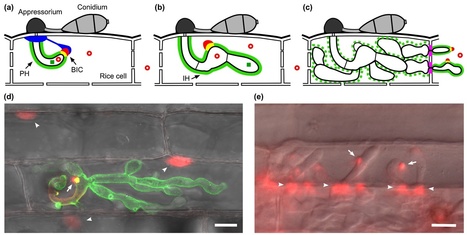
To cause riceblast disease, the fungus Magnaporthe oryzae produces biotrophic invasive hyphae that secrete effectors at the host–pathogen interface. Effectors facilitate disease development, but some (avirulence effectors) also trigger the host's resistance gene-mediated hypersensitive response and block disease. The number of cloned M. oryzae avirulence effector genes has recently doubled, largely based on resequencing with a Japanese field isolate and association of avirulence activity with presence/absence polymorphisms in novel genes for secreted proteins. Effectors secreted by hyphae in rice cells accumulate in biotrophic interfacial complexes, and this property correlates with their translocation across plasma membrane into the rice cytoplasm. Interestingly, the translocated effectors moved into surrounding uninvaded cells, suggesting that effectors prepare host cells before the fungus enters them.
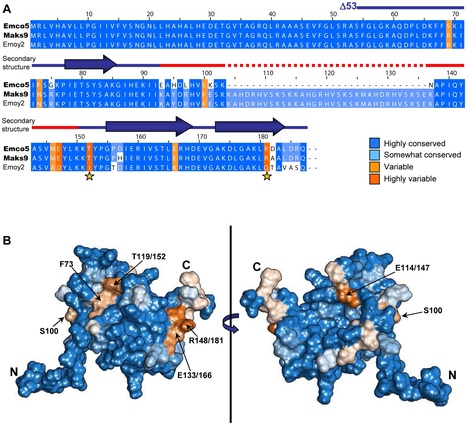
The oomycete Hyaloperonospora arabidopsidis (Hpa) is the causal agent of downy mildew on the model plant Arabidopsis thaliana and has been adapted as a model system to investigate pathogen virulence strategies and plant disease resistance mechanisms. Recognition of Hpa infection occurs when plant resistance proteins (R-genes) detect the presence or activity of pathogen-derived protein effectors delivered to the plant host. This study examines the Hpa effector ATR13 Emco5 and its recognition by RPP13-Nd, the cognate R-gene that triggers programmed cell death (HR) in the presence of recognized ATR13 variants. Herein, we use NMR to solve the backbone structure of ATR13 Emco5, revealing both a helical domain and a disordered internal loop. Additionally, we use site-directed and random mutagenesis to identify several amino acid residues involved in the recognition response conferred by RPP13-Nd. Using our structure as a scaffold, we map these residues to one of two surface-exposed patches of residues under diversifying selection. Exploring possible roles of the disordered region within the ATR13 structure, we perform domain swapping experiments and identify a peptide sequence involved in nucleolar localization. We conclude that ATR13 is a highly dynamic protein with no clear structural homologues that contains two surface-exposed patches of polymorphism, only one of which is involved in RPP13-Nd recognition specificity.
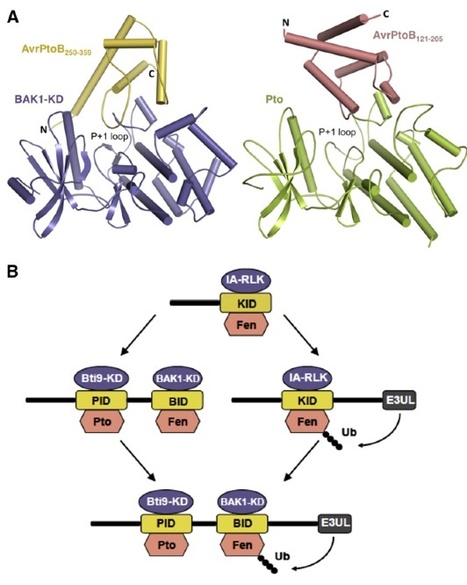
To infect plants, Pseudomonas syringae pv. tomato delivers ∼30 type III effector proteins into host cells, many of which interfere with PAMP-triggered immunity (PTI). One effector, AvrPtoB, suppresses PTI using a central domain to bind host BAK1, a kinase that acts with several pattern recognition receptors to activate defense signaling. A second AvrPtoB domain binds and suppresses the PTI-associated kinase Bti9 but is conversely recognized by the protein kinase Pto to activate effector-triggered immunity. We report the crystal structure of the AvrPtoB-BAK1 complex, which revealed structural similarity between these two AvrPtoB domains, suggesting that they arose by intragenic duplication. The BAK1 kinase domain is structurally similar to Pto, and a conserved region within both BAK1 and Pto interacts with AvrPtoB. BAK1 kinase activity is inhibited by AvrPtoB, and mutations at the interaction interface disrupt AvrPtoB virulence activity. These results shed light on a structural mechanism underlying host-pathogen coevolution.
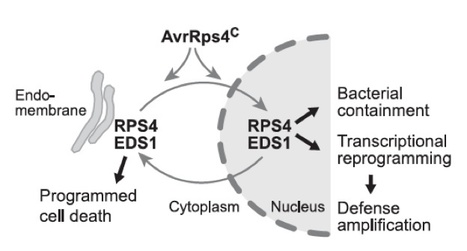
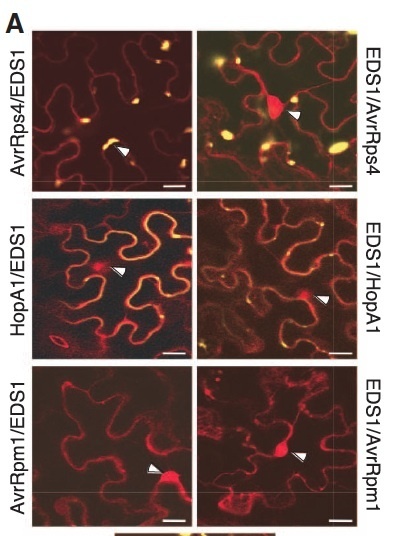
Pathogen effectors are intercepted by plant intracellular nucleotide binding–leucine-rich repeat (NB-LRR) receptors. However, processes linking receptor activation to downstream defenses remain obscure. Nucleo-cytoplasmic basal resistance regulator EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1) is indispensible for immunity mediated by TIR (Toll–interleukin-1 receptor)–NB-LRR receptors. We show that Arabidopsis EDS1 molecularly connects TIR-NB-LRR disease resistance protein RPS4 recognition of bacterial effector AvrRps4 to defense pathways. RPS4-EDS1 and AvrRps4-EDS1 complexes are detected inside nuclei of living tobacco cells after transient coexpression and in Arabidopsis soluble leaf extracts after resistance activation. Forced AvrRps4 localization to the host cytoplasm or nucleus reveals cell compartment–specific RPS4-EDS1 defense branches. Although nuclear processes restrict bacterial growth, programmed cell death and transcriptional resistance reinforcement require nucleo-cytoplasmic coordination. Thus, EDS1 behaves as an effector target and activated TIR-NB-LRR signal transducer for defenses across cell compartments.
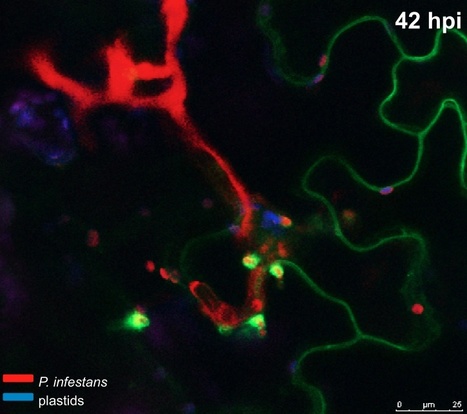
In response to pathogen attack, plant cells secrete antimicrobial molecules at the site of infection. However, how plant pathogens interfere with defense-related focal secretion remains poorly known. Here we show that the host-translocated RXLR-type effector protein AVRblb2 of the Irish potato famine pathogen Phytophthora infestans focally accumulates around haustoria, specialized infection structures that form inside plant cells, and promotes virulence by interfering with the execution of host defenses. AVRblb2 significantly enhances susceptibility of host plants to P. infestans by targeting the host papain-like cysteine protease C14 and specifically preventing its secretion into the apoplast. Plants altered in C14 expression were significantly affected in susceptibility to P. infestans in a manner consistent with a positive role of C14 in plant immunity. Our findings point to a unique counterdefense strategy that plant pathogens use to neutralize secreted host defense proteases. Effectors, such as AVRblb2, can be used as molecular probes to dissect focal immune responses at pathogen penetration sites.
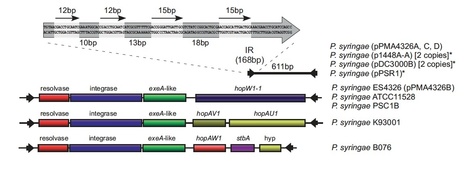
Miniature inverted terminal-repeat elements (MITEs) are non-autonomous mobile elements that have a significant impact on bacterial evolution. Here we characterize E622, a 611 bp virulence-associated MITE from Pseudomonas syringae, which contains no coding region, but has almost perfect 168 bp inverted repeats. Using an antibiotic-coupling assay, we show that E622 is transposable, and can mobilize an antibiotic resistance gene contained between its borders. Its predicted parent element, designated TnE622, has a typical transposon structure with a three-gene operon consisting of a resolvase, integrase, and exeA-like genes, which is bounded by the same terminal inverted repeats as E622. A broader genome-level survey of the E622/TnE622 inverted repeats identified homologs in Pseudomonas, Salmonella, Shewanella, Erwinia, Pantoea, and the cyanobacteria, Nostoc and Cyanothece, many of which appear to encompass known virulence genes, including toxins, enzymes, and type III secreted effectors. Its association with niche-specific genetic determinants, along with its persistence and evolutionary diversification indicates that this mobile element family has played a prominent role in the evolution of many agriculturally and clinically relevant pathogenic bacteria.
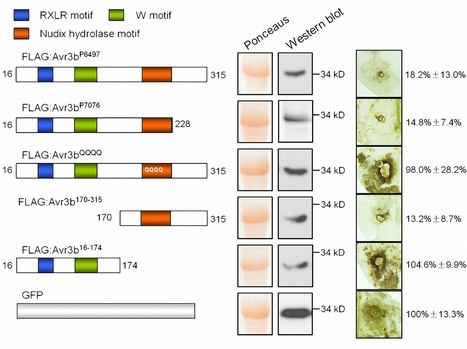
Plants have evolved pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) to protect themselves from infection by diverse pathogens. Avirulence (Avr) effectors that trigger plant ETI as a result of recognition by plant resistance (R) gene products have been identified in many plant pathogenic oomycetes and fungi. However, the virulence functions of oomycete and fungal Avr effectors remain largely unknown. Here, we combined bioinformatics and genetics to identify Avr3b, a new Avr gene from Phytophthora sojae, an oomycete pathogen that causes soybean root rot. Avr3b encodes a secreted protein with the RXLR host-targeting motif and C-terminal W and Nudix hydrolase motifs. Some isolates of P. sojae evade perception by the soybean R gene Rps3b through sequence mutation in Avr3b and lowered transcript accumulation. Transient expression of Avr3b in Nicotiana benthamiana increased susceptibility to P. capsici and P. parasitica, with significantly reduced accumulation of reactive oxygen species (ROS) around invasion sites. Biochemical assays confirmed that Avr3b is an ADP-ribose/NADH pyrophosphorylase, as predicted from the Nudix motif. Deletion of the Nudix motif of Avr3b abolished enzyme activity. Mutation of key residues in Nudix motif significantly impaired Avr3b virulence function but not the avirulence activity. Some Nudix hydrolases act as negative regulators of plant immunity, and thus Avr3b might be delivered into host cells as a Nudix hydrolase to impair host immunity. Avr3b homologues are present in several sequenced Phytophthora genomes, suggesting that Phytophthora pathogens might share similar strategies to suppress plant immunity.
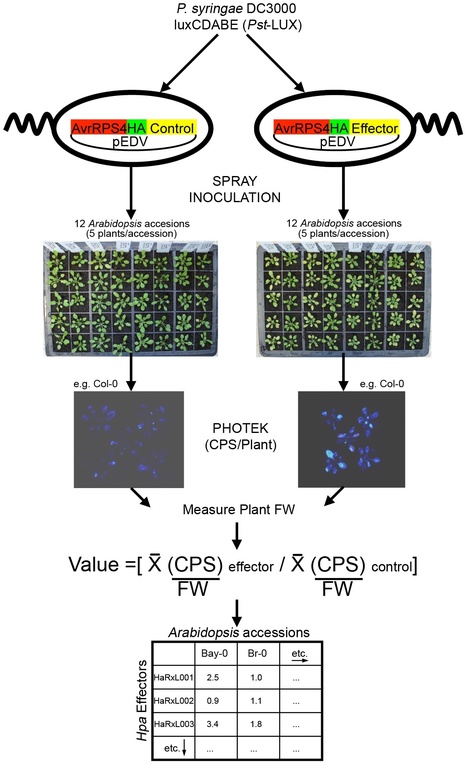
Oomycete pathogens cause diverse plant diseases. To successfully colonize their hosts, they deliver a suite of effector proteins that can attenuate plant defenses. In the oomycete downy mildews, effectors carry a signal peptide and an RxLR motif. Hyaloperonospora arabidopsidis (Hpa) causes downy mildew on the model plant Arabidopsis thaliana (Arabidopsis). We investigated if candidate effectors predicted in the genome sequence of Hpa isolate Emoy2 (HaRxLs) were able to manipulate host defenses in different Arabidopsis accessions. We developed a rapid and sensitive screening method to test HaRxLs by delivering them via the bacterial type-three secretion system (TTSS) of Pseudomonas syringae pv tomato DC3000-LUX (Pst-LUX) and assessing changes in Pst-LUX growth in planta on 12 Arabidopsis accessions. The majority (~70%) of the 64 candidates tested positively contributed to Pst-LUX growth on more than one accession indicating that Hpa virulence likely involves multiple effectors with weak accession-specific effects. Further screening with a Pst mutant (ΔCEL) showed that HaRxLs that allow enhanced Pst-LUX growth usually suppress callose deposition, a hallmark of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI). We found that HaRxLs are rarely strong avirulence determinants. Although some decreased Pst-LUX growth in particular accessions, none activated macroscopic cell death. Fewer HaRxLs conferred enhanced Pst growth on turnip, a non-host for Hpa, while several reduced it, consistent with the idea that turnip's non-host resistance against Hpa could involve a combination of recognized HaRxLs and ineffective HaRxLs. We verified our results by constitutively expressing in Arabidopsis a sub-set of HaRxLs. Several transgenic lines showed increased susceptibility to Hpa and attenuation of Arabidopsis PTI responses, confirming the HaRxLs' role in Hpa virulence. This study shows TTSS screening system provides a useful tool to test whether candidate effectors from eukaryotic pathogens can suppress/trigger plant defense mechanisms and to rank their effectiveness prior to subsequent mechanistic investigation.
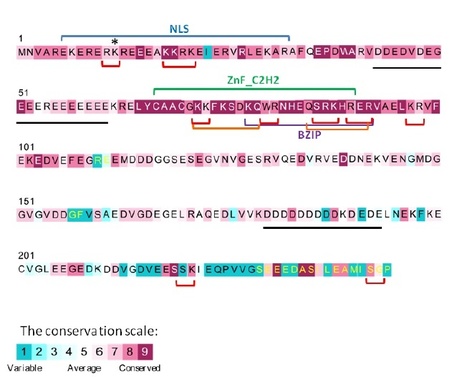
The type III effector HsvG of the gall-forming Pantoea agglomerans pv. gypsophilae (Pag) is a DNA-binding protein that is imported to the host nucleus and involved in host specificity. The DNA-binding region of HsvG was delineated to 266 amino acids located within a secondary structure region near the N-terminus of the protein, but did not display any homology to canonical DNA-binding motifs. A binding site selection procedure was used to isolate a target gene of HsvG, named HSVGT, in Gypsophila paniculata. HSVGT is a predicted acidic protein of the DnaJ family with 244 amino acids. It harbors characteristic conserved motifs of an eukaryotic transcription factor including a bipartite nuclear localization signal, zinc finger and leucine zipper DNA-binding motifs. Quantitative real-time PCR analysis demonstrated that HSVGT transcription is specifically induced in planta within 2 h after inoculation with the wild type Pag compared to hsvG mutant. Induction of HSVGT reached a peak of 6 fold at 4 h after inoculation and progressively declined thereafter. Gel-shift assay demonstrated that HsvG binds to the HSVGT promoter indicating that HSVGT is a direct target of HsvG. Our results support to the hypothesis that HsvG functions as a transcription factor in gypsophila.
Rice is atypical in that it is an agricultural cereal that is immune to fungal rust diseases. This report demonstrates that several cereal rust species (Puccinia graminis f. sp tritici, P. triticina, P. striiformis, and P. hordei) can infect rice and produce all the infection structures necessary for plant colonization, including specialized feeding cells (haustoria). Some rust infection sites are remarkably large and many plant cells are colonized, suggesting that nutrient uptake occurs to support this growth. Rice responds with an active, nonhost resistance (NHR) response that prevents fungal sporulation and that involves callose deposition, production of reactive oxygen species, and, occasionally, cell death. Genetic variation for the efficacy of NHR to wheat stem rust and wheat leaf rust was observed. Unlike cereal rusts, the rust pathogen (Melampsora lini) of the dicotyledenous plant flax (Linum usitatissimum) rarely successfully infects rice due to an apparent inability to recognize host-derived signals. Morphologically abnormal infection structures are produced and appressorial-like structures often don't coincide with stomata. These data suggest that basic compatibility is an important determinate of nonhost infection outcomes of rust diseases on cereals, with cereal rusts being more capable of infecting a cereal nonhost species compared with rust species that are adapted for dicot hosts.
|

|
Suggested by
dromius
|
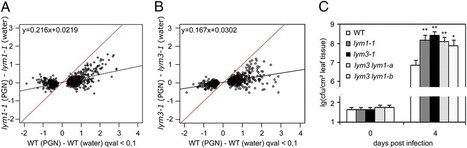
Recognition of microbial patterns by host pattern recognition receptors is a key step in immune activation in multicellular eukaryotes. Peptidoglycans (PGNs) are major components of bacterial cell walls that possess immunity-stimulating activities in metazoans and plants. Here we show that PGN sensing and immunity to bacterial infection in Arabidopsis thaliana requires three lysin-motif (LysM) domain proteins. LYM1 and LYM3 are plasma membrane proteins that physically interact with PGNs and mediate Arabidopsis sensitivity to structurally different PGNs from Gram-negative and Gram-positive bacteria. lym1 and lym3 mutants lack PGN-induced changes in transcriptome activity patterns, but respond to fungus-derived chitin, a pattern structurally related to PGNs, in a wild-type manner. Notably, lym1, lym3, and lym3 lym1 mutant genotypes exhibit supersusceptibility to infection with virulent Pseudomonas syringae pathovar tomato DC3000. Defects in basal immunity in lym3 lym1 double mutants resemble those observed in lym1 and lym3 single mutants, suggesting that both proteins are part of the same recognition system. We further show that deletion of CERK1, a LysM receptor kinase that had previously been implicated in chitin perception and immunity to fungal infection in Arabidopsis, phenocopies defects observed in lym1 and lym3 mutants, such as peptidoglycan insensitivity and enhanced susceptibility to bacterial infection. Altogether, our findings suggest that plants share with metazoans the ability to recognize bacterial PGNs. However, as Arabidopsis LysM domain proteins LYM1, LYM3, and CERK1 form a PGN recognition system that is unrelated to metazoan PGN receptors, we propose that lineage-specific PGN perception systems have arisen through convergent evolution.
DNA recognition by TAL effectors is mediated by tandem repeats, each 33 to 35 residues in length, that specify nucleotides via unique repeat variable diresidues (RVDs). The crystal structure of PthXo1 bound to its DNA target was determined using high-throughput computational structure prediction and validated by heavy-atom derivatization. Each repeat forms a left-handed, two-helix bundle that presents an RVD-containing loop to the DNA. The repeats self-associate to form a right-handed superhelix wrapped around the DNA major groove. The first RVD residue forms a stabilizing contact with the protein backbone, while the second makes a base-specific contact to the DNA sense strand. Two degenerate N-terminal repeats also interact with the DNA. Containing several RVDs and noncanonical associations, the structure illustrates the basis of TAL effector-DNA recognition.
Via dromius
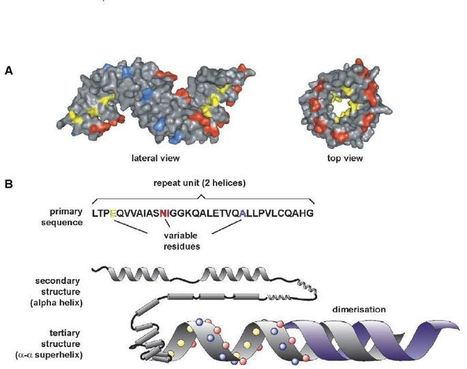
AvrBs3-like proteins are predicted to form a helical superstructure that resembles a tetratricopeptide repeat (TPR) fold. Variable repeat unit residues 4 (yellow), 12+13 (red) and 24 (blue) are depicted. (A) 3D Jury/MODELLER surface model was predicted based on the crystal structure of the TPR domain of the O-linked GLCNAC transferase (Protein Database (PDB) entry: 1w3b_A) and is displayed as lateral and top view. (B) Schematic illustration of the predicted right-handed α–α helical superstructure of AvrBs4 and its structural hierarchy. A second AvrBs3-like protein (blue) illustrates the postulated dimerization of this protein type.
Via dromius
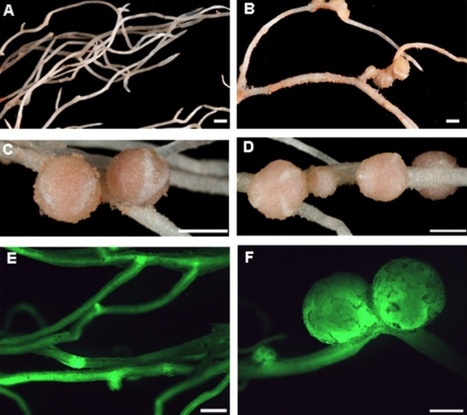
To allow rhizobial infection of legume roots, plant cell walls must be locally degraded for plant-made infection threads (ITs) to be formed. Here we identify a Lotus japonicus nodulation pectate lyase gene (LjNPL), which is induced in roots and root hairs by rhizobial nodulation (Nod) factors via activation of the nodulation signaling pathway and the NIN transcription factor. Two Ljnpl mutants produced uninfected nodules and most infections arrested as infection foci in root hairs or roots. The few partially infected nodules that did form contained large abnormal infections. The purified LjNPL protein had pectate lyase activity, demonstrating that this activity is required for rhizobia to penetrate the cell wall and initiate formation of plant-made infection threads. Therefore, we conclude that legume-determined degradation of plant cell walls is required for root infection during initiation of the symbiotic interaction between rhizobia and legumes.
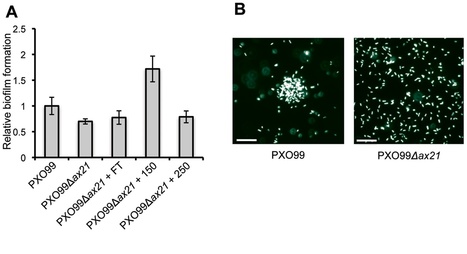
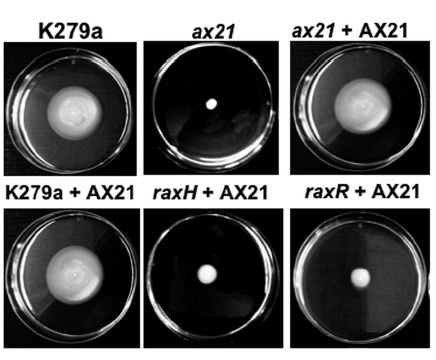
The rice XA21 pattern recognition receptor binds a type I secreted sulfated peptide, called axYS22, derived from the Ax21 (activator of XA21-mediated immunity) protein. The conservation of Ax21 in all sequenced Xanthomonas spp. and closely related genera suggests that Ax21 serves a key biological function. Here we show that the predicted N-terminal sequence of Ax21 is cleaved prior to secretion outside the cell and that mature Ax21 serves as a quorum sensing (QS) factor in Xanthomonas oryzae pv. oryzae. Ax21-mediated QS controls motility, biofilm formation and virulence. We provide genetic evidence that the Xoo RaxH histidine kinase serves as the bacterial receptor for Ax21. This work establishes a critical role for small protein-mediated QS in a Gram-negative bacterium.
Stenotrophomonas maltophilia encodes proteins related to the Rax proteins of Xanthomonas oryzae, which are required for the synthesis and secretion of the Ax21 protein. Here we show that Ax21 acts as a cell-cell signal to regulate a diverse range of functions, including virulence, in this nosocomial pathogen.
Plant resistance proteins detect the presence of specific pathogen effectors and initiate effector-triggered immunity. Few immune regulators downstream of resistance proteins have been identified, none of which are known virulence targets of effectors. We show that Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), a positive regulator of basal resistance and of effector-triggered immunity specifically mediated by Toll–interleukin-1 receptor–nucleotide binding–leucine-rich repeat (TIR-NB-LRR) resistance proteins, forms protein complexes with the TIR-NB-LRR disease resistance proteins RPS4 and RPS6 and with the negative immune regulator SRFR1 at a cytoplasmic membrane. Further, the cognate bacterial effectors AvrRps4 and HopA1 disrupt these EDS1 complexes. Tight association of EDS1 with TIR-NB-LRR–mediated immunity may therefore derive mainly from being guarded by TIR-NB-LRR proteins, and activation of this branch of effector-triggered immunity may directly connect to the basal resistance signaling pathway via EDS1.
Legume plants host nitrogen-fixing endosymbiotic Rhizobium bacteria in root nodules. In Medicago truncatula, the bacteria undergo an irreversible (terminal) differentiation mediated by hitherto unidentified plant factors. We demonstrated that these factors are nodule-specific cysteine-rich (NCR) peptides that are targeted to the bacteria and enter the bacterial membrane and cytosol. Obstruction of NCR transport in the dnf1-1 signal peptidase mutant correlated with the absence of terminal bacterial differentiation. On the contrary, ectopic expression of NCRs in legumes devoid of NCRs or challenge of cultured rhizobia with peptides provoked symptoms of terminal differentiation. Because NCRs resemble antimicrobial peptides, our findings reveal a previously unknown innovation of the host plant, which adopts effectors of the innate immune system for symbiosis to manipulate the cell fate of endosymbiotic bacteria.
It's been dubbed the foot and mouth of the tree world. Phytophthora ramorum or sudden oak death as its commonly known is ravaging forests across the UK resulting in millions of trees being cut down. The disease has spread from the South West to Wales, the peaks and even as far north as the Isle of Mull. But experts say they are finding fewer and fewer new outbreaks. Today on Open Country, Helen Mark visits The South West, the region that's hardest hit, to find out what impact this disease is continuing to have on the countryside and whether there are signs that we are finally getting on top of it.
Phytoplasmas are insect-transmitted phytopathogenic bacteria that can alter plant morphology and the longevity and reproduction rates and behavior of their insect vectors. There are various examples of animal and plant parasites that alter the host phenotype to attract insect vectors, but it is unclear how these parasites accomplish this. We hypothesized that phytoplasmas produce effectors that modulate specific targets in their hosts leading to the changes in plant development and insect performance. Previously, we sequenced and mined the genome of Aster Yellows phytoplasma strain Witches’ Broom (AY-WB) and identified 56 candidate effectors. Here, we report that the secreted AY-WB protein 11 (SAP11) effector modulates plant defense responses to the advantage of the AY-WB insect vector Macrosteles quadrilineatus. SAP11 binds and destabilizes Arabidopsis CINCINNATA (CIN)-related TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTORS 1 and 2 (TCP) transcription factors, which control plant development and promote the expression of lipoxygenase (LOX) genes involved in jasmonate (JA) synthesis. Both the Arabidopsis SAP11 lines and AY-WB–infected plants produce less JA on wounding. Furthermore, the AY-WB insect vector produces more offspring on AY-WB–infected plants, SAP11 transgenic lines, and plants impaired in CIN-TCP and JA synthesis. Thus, SAP11-mediated destabilization of CIN-TCPs leads to the down-regulation of LOX2 expression and JA synthesis and an increase in M. quadrilineatus progeny. Phytoplasmas are obligate inhabitants of their plant host and insect vectors, in which the latter transmits AY-WB to a diverse range of plant species. This finding demonstrates that pathogen effectors can reach beyond the pathogen–host interface to modulate a third organism in the biological interaction.
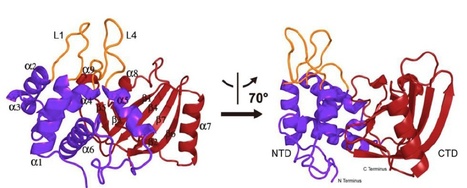
The Pseudomonas syringae type III effector HopU1 is a mono-ADP-ribosyltransferase (mADP-RT) that is injected into plant cells by the type III protein secretion system. Inside the plant cell it suppresses immunity by modifying RNA-binding proteins including the glycine-rich RNA-binding protein GRP7. The crystal structure of HopU1 at 2.7 angstrom resolution reveals two unique protruding loops, L1 and L4 not found in other mADP-RTs. Site-directed mutagenesis demonstrates these loops are essential for substrate recognition and enzymatic activity. HopU1 ADP-ribosylates the conserved arginine 49 (R49) of GRP7 and this reduces GRP7's ability to bind RNA in vitro. In vivo, expression of GRP7 with R49 replaced with lysine does not complement the reduced immune responses of the Arabidopsis thaliana grp7-1 mutant demonstrating the importance of this residue for GRP7 function. These data provide mechanistic details how HopU1 recognizes this novel type of substrate and highlights the role of GRP7 in plant immunity.
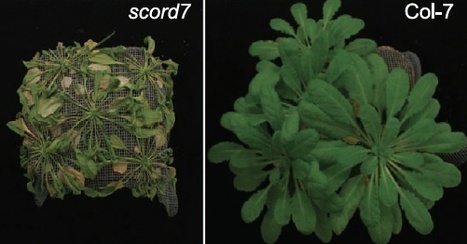
Bacterial infection of plants often begins with colonization of the plant surface, followed by entry into the plant through wounds and natural openings (such as stomata), multiplication in the intercellular space (apoplast) of the infected tissues, and dissemination of bacteria to other plants. Historically, most studies assess bacterial infection based on final outcomes of disease and/or pathogen growth using whole infected tissues; few studies have genetically distinguished the contribution of different host cell types in response to an infection. The phytotoxin coronatine (COR) is produced by several pathovars of Pseudomonas syringae. COR-deficient mutants of P. s. tomato (Pst) DC3000 are severely compromised in virulence, especially when inoculated onto the plant surface. We report here a genetic screen to identify Arabidopsis mutants that could rescue the virulence of COR-deficient mutant bacteria. Among the susceptible to coronatine-deficient Pst DC3000 (scord) mutants were two that were defective in stomatal closure response, two that were defective in apoplast defense, and four that were defective in both stomatal and apoplast defense. Isolation of these three classes of mutants suggests that stomatal and apoplastic defenses are integrated in plants, but are genetically separable, and that COR is important for Pst DC3000 to overcome both stomatal guard cell- and apoplastic mesophyll cell-based defenses. Of the six mutants defective in bacterium-triggered stomatal closure, three are defective in salicylic acid (SA)-induced stomatal closure, but exhibit normal stomatal closure in response to abscisic acid (ABA), and scord7 is compromised in both SA- and ABA-induced stomatal closure. We have cloned SCORD3, which is required for salicylic acid (SA) biosynthesis, and SCORD5, which encodes an ATP-binding cassette (ABC) protein, AtGCN20/AtABCF3, predicted to be involved in stress-associated protein translation control. Identification of SCORD5 begins to implicate an important role of stress-associated protein translation in stomatal guard cell signaling in response to microbe-associated molecular patterns and bacterial infection.
|
 Your new post is loading...
Your new post is loading...
 Your new post is loading...
Your new post is loading...