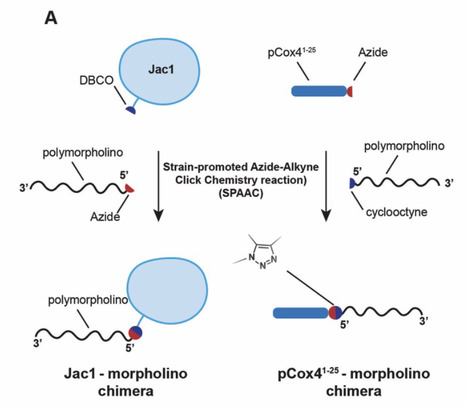
Cooperative catalysis, in which multiple catalytic units operate synergistically, underpins a variety of synthetically and mechanistically important organic reactions1–4. Despite its potential utility in new reactivity contexts, approaches to the discovery of cooperative catalysts have been limited, typically relying on serendipity or on prior knowledge of single-catalyst reactivity1,5. Systematic searches for unanticipated types of catalyst cooperativity must contend with vast combinatorial complexity and are therefore not undertaken6–10. Here, we describe a pooling–deconvolution algorithm, inspired by group testing11, that identifies cooperative catalyst behaviors with low experimental cost while accommodating potential inhibitory effects between catalyst candidates. The workflow was validated first on simulated cooperativity data, and then by experimentally identifying previously documented cooperativity between organocatalysts in an enantioselective oxetane-opening reaction. The workflow was then applied in a discovery context to a Pd-catalyzed decarbonylative cross-coupling reaction, enabling the identification of several ligand pairs that promote the target transformation at substantially lower catalyst loading and temperature than previously reported with single ligand systems.
Follow, research and publish the best content
Get Started for FREE
Sign up with Facebook Sign up with X
I don't have a Facebook or a X account
Already have an account: Login
 Your new post is loading... Your new post is loading...
 Your new post is loading... Your new post is loading...
Marcus Jorelle's curator insight,
December 13, 2024 6:08 AM
Sierra MatchKing Bullets 22 Caliber (224 Diameter) 69 Grain Hollow Point Boat Tail: The Ultimate Choice for Precision Shooting ▎Introduction When it comes to precision shooting, every detail matters—from the rifle to the ammunition. Among the top choices for competitive shooters and enthusiasts alike are the Sierra MatchKing Bullets 22 Caliber (224 Diameter) 69 Grain Hollow Point Boat Tail. Renowned for their exceptional accuracy and performance, these bullets are designed to meet the rigorous demands of long-range shooting. In this article, we will explore the features, benefits, and applications of these premium bullets while incorporating essential SEO keywords to enhance your searchability. ▎What Are Sierra MatchKing Bullets? Sierra MatchKing bullets are specifically engineered for competitive shooting and precision applications. With a focus on accuracy, these bullets are crafted with meticulous attention to detail, ensuring they perform consistently in various conditions. The 22 caliber (224 diameter) designation makes them suitable for a range of rifles, while the 69 grain weight strikes a balance between velocity and stability. ▎Key Features of Sierra MatchKing Bullets 1. Hollow Point Design: The hollow point design enhances ballistic performance by promoting controlled expansion upon impact. This feature is particularly beneficial for target shooting, as it helps maintain accuracy at longer distances. 2. Boat Tail Configuration: The boat tail design reduces drag and improves aerodynamics, allowing for flatter trajectories and better stability in flight. This results in enhanced accuracy and reduced wind drift—critical factors for long-range shooters. 3. High-Quality Manufacturing: Sierra is known for its commitment to quality, and the MatchKing bullets are no exception. Each bullet undergoes rigorous testing to ensure consistency in weight, shape, and performance. ▎Benefits of Using Sierra MatchKing Bullets ▎1. Exceptional Accuracy The primary advantage of using Sierra MatchKing bullets is their unmatched accuracy. Whether you are participating in a competition or honing your skills at the range, these bullets are designed to deliver tight groupings, giving you confidence in your shots. ▎2. Versatility While primarily designed for match shooting, the 22 caliber 69 grain hollow point boat tail bullets can also be utilized for varmint hunting and other applications where precision is paramount. Their versatility makes them a valuable addition to any shooter’s arsenal. ▎3. Consistent Performance Thanks to their high-quality construction, MatchKing bullets provide consistent performance across various shooting conditions. This reliability ensures that you can trust your ammunition when it matters most. ▎Applications of Sierra MatchKing Bullets Sierra MatchKing bullets are ideal for: • Competitive Shooting: Perfect for benchrest competitions, high-power matches, and other precision shooting events. • Target Practice: Excellent choice for those looking to improve their marksmanship skills. • Varmint Hunting: Effective for taking down small game due to their accurate and controlled expansion. ▎How to Load Sierra MatchKing Bullets Loading Sierra MatchKing bullets requires careful attention to detail to maximize their potential. Here are some tips: 1. Choose the Right Powder: Select a powder that complements the bullet weight and your specific rifle. Refer to reloading manuals for recommended powder types and charges. 2. Set Proper OAL (Overall Length): Adjust the seating depth to achieve the optimal overall length for your specific firearm. This can significantly affect accuracy. 3. Test Different Loads: Experiment with various loads to find the combination that delivers the best performance in your rifle.https://canada-reloading.shop/product/federal-firestic…-powder-10-count/ https://canada-reloading.shop/product/fiocchi-primers-…09-w-type-1000ct/ https://canada-reloading.shop/product/fiocchi-small-pi…l-magnum-primers/ https://canada-reloading.shop/product/large-rifle-magn…formance-primers/ https://canada-reloading.shop/product/pmc-small-pistol…rimers-two-trays/ https://canada-reloading.shop/product/remington-10-percussion-caps-100/ https://canada-reloading.shop/product/remington-11-percussion-caps/ https://canada-reloading.shop/product/remington-209ml-…ader-primers-100/ https://canada-reloading.shop/product/remington-22617-…-caps-100-ct-tin/ https://canada-reloading.shop/product/rws-1075-plus-percussion-caps-11/ https://canada-reloading.shop/product/rws-4-flange-200-ct-musket-caps/ https://canada-reloading.shop/product/rws-11-percussio…250-to-1000-pack/ https://canada-reloading.shop/product/rws-musket-caps-…200-to-5000-pack/ https://canada-reloading.shop/product/schuetzen-musket…00-to-5000-count/ https://canada-reloading.shop/product/small-rifle-magn…formance-primers/ https://canada-reloading.shop/product/tulammo-large-ri…imers-1000-count/ https://canada-reloading.shop/product/tulammo-small-ri…imers-1000-count/ https://canada-reloading.shop/product/winchester-percussion-caps-11-100/ https://canada-reloading.shop/product/209-shotshell-wo…formance-primers/ https://canada-reloading.shop/product/cci-percussion-c…0-10-cans-of-100/ https://canada-reloading.shop/product/cci-primers-209-…-10-trays-of-100/ https://canada-reloading.shop/product/cci-primers-209m…-10-trays-of-100/ https://canada-reloading.shop/product/cheddite-shotshell-primer-209/ https://canada-reloading.shop/product/federal-209a-shotshell-primers/ https://canada-reloading.shop/product/federal-primers-…-10-trays-of-100/ https://canada-reloading.shop/product/fiocchi-primers-…-10-trays-of-100/ https://canada-reloading.shop/product/nobel-sport-shot…-10000-pack-case/ https://canada-reloading.shop/product/nobelsport-prime…-50-trays-of-100/ https://canada-reloading.shop/product/remington-premie…rs-209-shotshell/ https://canada-reloading.shop/product/rio-209-g-1000-shotshell-primers/ https://canada-reloading.shop/product/winchester-prime…-10-trays-of-100/ https://canada-reloading.shop/product/fort-smith-fsaap…ol-boxer-primers/ https://canada-reloading.shop/product/cci-small-pistol…-10-trays-of-100/ https://canada-reloading.shop/product/cci-small-pistol…-10-trays-of-100/ https://canada-reloading.shop/product/federal-premium-…-10-trays-of-100/ https://canada-reloading.shop/product/federal-premium-…-10-trays-of-100/ https://canada-reloading.shop/product/federal-small-pi…-10-trays-of-100/ https://canada-reloading.shop/product/federal-small-pi…-10-trays-of-100/ https://canada-reloading.shop/product/remington-small-…-10-trays-of-100/ https://canada-reloading.shop/product/remington-small-…-10-trays-of-100/ https://canada-reloading.shop/product/small-pistol-wol…formance-primers/ https://canada-reloading.shop/product/small-pistol-mag…formance-primers/ https://canada-reloading.shop/product/winchester-small…-10-trays-of-100/ https://canada-reloading.shop/product/winchester-small…-10-trays-of-100/ https://canada-reloading.shop/product/winchester-small…-10-trays-of-100/ https://canada-reloading.shop/product/winchester-usa-r…-10-trays-of-100/ https://canada-reloading.shop/product/fort-smith-fsaap…le-boxer-primers/ https://canada-reloading.shop/product/cci-small-rifle-…-10-trays-of-100/ https://canada-reloading.shop/product/cci-small-rifle-…-10-trays-of-100/ https://canada-reloading.shop/product/cci-small-rifle-…-10-trays-of-100/ https://canada-reloading.shop/product/cci-small-rifle-…-10-trays-of-100/ https://canada-reloading.shop/product/federal-premium-…-10-trays-of-100/ https://canada-reloading.shop/product/federal-small-ri…-10-trays-of-100/ https://canada-reloading.shop/product/magtech-small-ri…imers-1000-count/ https://canada-reloading.shop/product/pmc-small-rifle-magnum-primers/ https://canada-reloading.shop/product/remington-small-…-10-trays-of-100/ https://canada-reloading.shop/product/remington-small-…-10-trays-of-100/ https://canada-reloading.shop/product/hornady-brass-45…ess-3″-box-of-20/ https://canada-reloading.shop/product/hornady-brass-6-5-prc-box-of-50/ https://canada-reloading.shop/product/lapua-brass-30-0…field-box-of-100/ https://canada-reloading.shop/product/lapua-brass-338-…agnum-box-of-100/ https://canada-reloading.shop/product/lapua-brass-6-5-creedmoor/ https://canada-reloading.shop/product/norma-brass-shoo…magnum-box-of-50/ https://canada-reloading.shop/product/nosler-brass-6-5…dmoor-bag-of-100/ https://canada-reloading.shop/product/nosler-custom-br…osler-box-of-100/ https://canada-reloading.shop/product/nosler-custom-br…hester-box-of-50/ https://canada-reloading.shop/product/nosler-custom-br…oulder-box-of-50/ https://canada-reloading.shop/product/nosler-custom-br…magnum-box-of-25/ https://canada-reloading.shop/product/nosler-custom-co…-point-boat-tail/ https://canada-reloading.shop/product/once-fired-brass…00-bulk-packaged/ https://canada-reloading.shop/product/sierra-matchking…-point-boat-tail/ https://canada-reloading.shop/product/sig-sauer-brass-…ington-bag-of-50/ https://canada-reloading.shop/product/starline-brass-223-remington/ https://canada-reloading.shop/product/starline-brass-357-sig/ https://canada-reloading.shop/product/starline-brass-38-55-wcf-2-082″/ https://canada-reloading.shop/product/starline-brass-45-acp/ https://canada-reloading.shop/product/starline-brass-45-auto-rim-not-acp/ https://canada-reloading.shop/product/starline-brass-458-socom/ https://canada-reloading.shop/product/starline-brass-5-56x45mm-nato/ https://canada-reloading.shop/product/top-brass-premiu…300-aac-blackout/ https://canada-reloading.shop/product/top-brass-premiu…ired-brass-40-sw/ https://canada-reloading.shop/product/hodgdon-2024-ann…reloading-manual/ https://canada-reloading.shop/product/hornady-11th-edi…reloading-manual/ https://canada-reloading.shop/product/loadbooks-usa-12…reloading-manual/ https://canada-reloading.shop/product/lyman-pistol-rev…book-3rd-edition/ https://canada-reloading.shop/product/norma-reloading-manual-2nd-edition/ https://canada-reloading.shop/product/nosler-reloading-manual-9/ https://canada-reloading.shop/product/sierra-6th-editi…reloading-manual/ https://canada-reloading.shop/product/speer-reloading-manual-15/ https://canada-reloading.shop/product/swift-reloading-manual-2/ https://canada-reloading.shop/product/tom-rosters-hevi…nual-2nd-edition/ https://canada-reloading.shop/product/top-brass-premiu…ired-brass-40-sw/ https://canada-reloading.shop/product/federal-premium-…-point-boat-tail/ https://canada-reloading.shop/product/hornady-superfor…-v-max-box-of-20/ "Referring page title" "Referring page URL" "Language" "Platform" "Referring page HTTP code" "Domain rating" "UR" "Domain traffic" "Referring domains" "Linked domains" "External links" "Page traffic" "Keywords" "Target URL" "Left context" "Anchor" "Right context" "Redirect Chain URLs" "Redirect Chain status codes" "Type" "Content" "Nofollow" "UGC" "Sponsored" "Rendered" "Raw" "Lost status" "Drop reason" "Discovered status" "First seen" "Last seen" "Lost" "Author" "Links in group" "YouTube Creator Blog [UK]: Ride to Success with 63fixedtv, September’s Rising Partner" "https://youtubecreator-uk.googleblog.com/2012/09/ride-to-success-with-63fixedtv.html" "en" "" "200" "94" "5" "210429" "29" "180" "542" "0" "0" "http://ssgunstore.com/" "" "Buy sig sauer online" "" "" "" "text" "false" "true" "false" "false" "true" "true" "" "" "pagefound" "2022-07-11 14:53:04" "2024-05-29 07:01:22" "" "" "2" "沖縄対策本部■沖縄に現れた若き愛国ヒロインのスッキリする名スピーチ - 沖縄対策本部" "https://blog.goo.ne.jp/jiritsukokka/e/55ac732a803559c65c0865560848bb22" "" "" "200" "90" "6" "24746634" "22" "711" "1879" "0" "0" "https://ssgunstore.com/" "(" "Sig Sauer" ")" "" "" "text" "false" "true" "false" "false" "false" "true" "" "" "linkfound" "2021-07-09 02:55:40" "2024-05-29 17:13:03" "" "" "1" "Immagine Immagina | Germaine Muller" "http://www.germainemuller.altervista.org/?page_id=172" "it" "wordpress" "200" "90" "0.6" "2411712" "93" "5554" "13863" "0" "0" "https://ssgunstore.com/" "" "sig sauer sp2022" "" "" "" "text" "false" "true" "false" "false" "true" "true" "" "" "linkrestored" "2022-07-04 22:49:07" "2024-05-27 21:28:57" "" "" "1" "Up Against the Wall: How to Break Up Your Day so You Feel Great – Magellan Chiropractic" "http://jonesborochiropractor.flywheelsites.com/up-against-the-wall-how-to-break-up-your-day-so-you-feel-great/" "" "wordpress" "200" "88" "1.1" "15265" "2" "3840" "8378" "0" "0" "https://ssgunstore.com/" "" "sig sauer ar 15" "" "" "" "text" "false" "true" "true" "false" "true" "true" "" "" "pagefound" "2023-05-27 08:35:04" "2024-05-27 18:49:19" "" "" "2" "The Moo's News » Blog Archive » Click To Tweet" "http://fatcow.com/blog/?p=1137" "en" "ecommerce, wordpress" "200" "86" "19" "16248" "1158" "13023" "30474" "0" "6" "https://ssgunstore.com/" "" "sig sauer p226" "Says:" "" "" "text" "false" "true" "false" "false" "false" "true" "" "" "linkrestored" "2021-03-31 08:20:02" "2024-05-29 20:22:08" "" "" "2" "EINMANNDING – Kommentare zu Heute , Morgentext vom 28.06. 2017" "http://wordpress.p255834.webspaceconfig.de/?comments_popup=5817" "de" "" "200" "85" "9" "5551" "290" "12307" "21416" "0" "0" "https://ssgunstore.com/" "Kommentar von" "Trevor Mackey" "— 5. April 2021 zu 1:33" "" "" "text" "false" "true" "false" "false" "false" "true" "" "" "pagefound" "2024-01-05 06:21:59" "2024-04-30 15:24:48" "" "" "3" "maquete_2 – Adaptpolis" "http://adaptpolis.fa.ulisboa.pt/?attachment_id=334" "" "wordpress" "200" "82" "0.1" "273078" "111" "7539" "11901" "0" "0" "https://ssgunstore.com/" "" "sig sauer rattler" "" "" "" "text" "false" "true" "true" "false" "false" "true" "" "" "pagefound" "2023-01-06 19:43:36" "2024-05-29 15:48:06" "" "" "1" "What might a post-Wonga world look like? - University of Birmingham" "https://www.birminghamdev.bham.ac.uk/research/perspective/what-might-a-post-wonga-world-look-like" "en" "" "200" "80" "0" "29063" "0" "3994" "13855" "0" "0" "https://ssgunstore.com/" "At 5:25AM on 21 July 2021," "sig sauer guns shop" "wrote" "" "" "text" "false" "true" "false" "false" "false" "true" "" "" "pagefound" "2024-03-27 10:10:12" "2024-04-29 05:20:04" "" "" "1" "Baseball Talk » Blog Archive » Hamilton and Rangers agree to deal" "https://blogs.bgsu.edu/mlblog/2011/02/10/hamilton-and-rangers-agree-to-deal/" "en" "wordpress" "200" "80" "4.3" "234573" "141" "6158" "12700" "0" "4" "https://ssgunstore.com/" "to create this actual post amazing. Excellent process!My page ::" "sig sauer p228" "" "" "" "text" "false" "true" "true" "false" "false" "true" "" "" "linkfound" "2022-06-29 18:25:00" "2024-05-30 01:38:43" "" "" "1" "How Is A Man Freed After 23 Years? – Juris Magazine" "https://sites.law.duq.edu/juris/2013/03/27/how-is-a-man-freed-after-23-years/" "" "wordpress" "200" "78" "3.1" "110203" "47" "7446" "13671" "0" "3" "https://ssgunstore.com/" "" "Mammie Maguire" "" "" "" "text" "false" "true" "true" "false" "false" "true" "" "" "linkfound" "2023-11-04 16:44:53" "2024-05-29 08:38:20" "" "" "1" "buy ayahuasca - On Feet Nation" "https://www.onfeetnation.com/profiles/blogs/buy-ayahuasca-1?xg_source=activity" "en" "" "200" "77" "3.8" "8" "0" "23" "34" "0" "0" "https://ssgunstore.com/" "" "sig sauer, sig sauer p320, sig sauer p365, sig sauer p226, sig saue..." "" "http://ssgunstore.com/" "301" "text" "false" "true" "false" "false" "false" "true" "" "" "linkrestored" "2021-10-12 17:19:30" "2024-05-15 03:29:06" "" "" "8" "For the Love of Food: Get Me to the Greek" "http://healthyeating.sunnybrook.ca/2013/02/get-me-to-greek.html" "en" "" "200" "75" "4.6" "68539" "24" "217" "528" "0" "0" "https://ssgunstore.com/" "" "Firearms For sale, sig sauer p320 compact, sig sauer sp2022, sig sauer p320 for sale, sig sauer p365 for sale, sig sauer p250, sig sauer 365, sig sauer tread, sig sauer p226 legion, sig sauer mcx virtus, sig sauer p320 x5, sig sauer 380, sig sauer p210, sig sauer legion, sig sauer p320 m17, sig sauer pistols, sig sauer p320 x5 legion, sig sauer p228, sig sauer p226 price, sig sauer p365 price, sig ..." "" "" "" "text" "false" "true" "false" "false" "true" "true" "" "" "linkfound" "2023-07-06 15:43:32" "2024-05-28 15:57:43" "" "" "1" "EDDL 5141 – Benefits and Challenges Activity – sdeighton's Portfolio" "https://sdeighton-portfolio.eddl.tru.ca/2019/01/22/eddl-5141-benefits-and-challenges-activity/" "en" "wordpress" "200" "74" "4.4" "108624" "47" "4166" "7570" "0" "0" "https://ssgunstore.com/" "" "sig sauer m18" "" "" "" "text" "false" "true" "true" "false" "false" "true" "" "" "linkfound" "2022-06-30 18:15:14" "2024-05-28 23:46:27" "" "" "2" "31 companies participate in AIT Career Fair – AIT Research Portal" "https://research.ait.ac.th/2018/03/14/31-companies-participate-in-ait-career-fair/" "en" "wordpress" "200" "73" "4.2" "49074" "51" "7473" "12596" "0" "0" "https://ssgunstore.com/" "" "sig sauer tread" "" "" "" "text" "false" "true" "true" "false" "false" "true" "" "" "linkrestored" "2023-10-31 15:56:15" "2024-05-29 09:57:32" "" "" "2" "Majelis Ta’lim Uswatun Nisa Peringati Maulid - BIRO ADMINISTRASI PIMPINAN SETDA ACEH" "https://humas.acehprov.go.id/majelis-talim-uswatun-nisa-peringati-maulid/" "id, en" "ecommerce, wordpress" "200" "71" "1.8" "629960" "35" "2169" "3365" "0" "2" "https://ssgunstore.com/" "" "sig sauer p229" "" "" "" "text" "false" "true" "true" "false" "true" "true" "" "" "linkrestored" "2022-12-22 11:51:32" "2024-05-13 05:12:57" "" "" "1" "Rio Anhanduí pede socorro - Hidrologia, Erosão e Sedimentos - HEroS" "https://heros.ufms.br/rio-anhandui-pede-socorro/" "pt" "wordpress" "200" "70" "4.5" "123300" "79" "8555" "14327" "0" "1" "https://ssgunstore.com/" "" "sig sauer p365" "" "" "" "text" "false" "true" "true" "false" "true" "true" "" "" "linkrestored" "2021-04-07 22:14:28" "2024-05-28 21:29:27" "" "Pedro Zamboni" "2" "Olá, mundo! | IFA"
|
|