Your new post is loading...
 Your new post is loading...

|
Scooped by
nrip
May 15, 2021 12:00 PM
|
COVID-19 Research Resources to develop your own study materials
The Covid-19 Research Implementation and Knowledge Hub of the Global Health Network contains a wide range of resources to guide teams in developing their own study materials and support high-quality, rapid implementation of critical COVID-19 studies in various settings. The Global Health Network is running this crowdsourcing initiative to support a faster implementation of research studies during this pandemic. It slow and unnecessary for each group wanting to set up a study to start from scratch; when there are excellent studies already running. The Hub is being continually updated so its good to bookmark it so you can check it regularly for new resources that could help your research team with your COVID-19 study. Link to the Covid-19 Research Implementation and Knowledge Hub

|
Scooped by
nrip
January 16, 2025 10:25 PM
|
In March 2022, a concerning rise in cases of unexplained paediatric hepatitis was reported in multiple countries. Cases were defined as acute hepatitis with serum transaminases >500 U/L (aspartate transaminase (AST) or alanine transaminase (ALT)) in children aged 16 or under. In this study the authors explore a simple federated data analytics method to search for evidence of unreported cases using routinely held data. They conducted a pragmatic survey to analyse changes in the proportion of hospitalised children with elevated AST or ALT over time. In addition, they studied the feasibility of using routinely collected clinical laboratory results to detect or follow up an outbreak of an infectious disease. Results: Our approach of sharing a simple software tool for local use enabled rapid, federated data analysis. 34 hospitals in the UK, the Netherlands, Ireland and Curaçao were asked to contribute summary data, and 30 (88%) submitted their data. For all locations combined, the rate of elevated AST or ALT measurements in the period of interest was not elevated (Z-score -0.46; p 0.64). Results from individual regions were discordant, with a higher rate of elevated AST or ALT values in the Netherlands (Z-score 4.48; p<0.0001), driven by results from a single centre in Utrecht. We did not observe any clear indication of changes in primary care activity or test results in the same period. Conclusions: Hospital laboratories collect large amounts of data on a daily basis that can potentially be of use for disease surveillance, but these are currently not optimally utilised. Federated analytics using non-disclosive, summary-level laboratory data sharing was successful, safe and efficient. The approach holds potential as a tool for pandemic surveillance in future outbreaks. Our findings do not indicate the presence of a broader outbreak of mild hepatitis cases among young children, although there was an increase in elevated AST or ALT values locally in the Netherlands. read the original unedited research work at https://publichealth.jmir.org/2024/1/e55376/

|
Scooped by
nrip
October 16, 2024 11:24 PM
|
During the peak of the winter 2020-2021 surge, the number of weekly reported COVID-19 outbreaks in Washington State was 231; the majority occurred in high-priority settings such as workplaces, community settings, and schools. The Washington State Department of Health used automated address matching to identify clusters at health care facilities. No other systematic, statewide outbreak detection methods were in place. This was a gap given the high volume of cases, which delayed investigations and decreased data completeness, potentially leading to undetected outbreaks. We initiated statewide cluster detection using SaTScan, implementing a space-time permutation model to identify COVID-19 clusters for investigation.
Objective: To improve outbreak detection, the Washington State Department of Health initiated a systematic cluster detection model to identify timely and actionable COVID-19 clusters for local health jurisdiction (LHJ) investigation and resource prioritization.
This report details the model’s implementation and the assessment of the tool’s effectiveness. Regardless of population or incidence, the model identified reasonably sized, timely clusters statewide, meeting the objective. Among some high-priority settings subject to public health interventions throughout the pandemic, such as schools and community settings, the model identified clusters that were matched to reported outbreaks. In workplaces, another high-priority setting, results suggest the model might be able to identify outbreaks sooner than existing outbreak detection methods.
read the research paper at https://publichealth.jmir.org/2024/1/e49871/

|
Scooped by
nrip
October 7, 2024 1:14 PM
|
Trust is fundamental to cooperation, essential in times of crisis. Researching and understanding trust networks and perceptions of trustworthiness is therefore crucial in preparing for future health shocks, write Heidi Larson and colleagues -
Although the longstanding public trust in healthcare professionals still holds, trust is waning among healthcare professionals themselves and needs further research -
Implementation science that evaluates setting specific approaches to building trust both before and in the context of crises is needed -
Beyond studies on dyadic trust relations, networks of trust need investigation to inform strategies for different settings as a key part of preparedness for future health shocks -
To make progress in research on trust, we need a dynamic systems approach, particularly crucial in the context of a health shock with its evolving uncertainties and multiple effects across the system and over time read more at https://www.bmj.com/content/387/bmj-2023-078464.short?rss=1

|
Rescooped by
nrip
from healthcare technology
May 7, 2024 10:27 PM
|
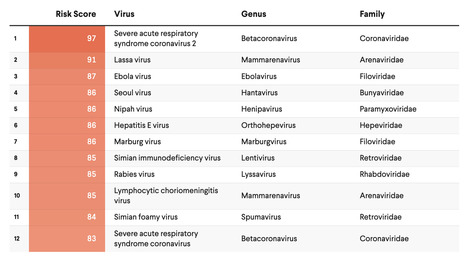
The past several decades have seen an alarming spike in communicable disease outbreaks worldwide. Given a confluence of host, virologic, environmental, and human factors, experts agree that the next pandemic could already be on the horizon. In a globalized world, changes in how people use land and interact with their ecosystems—such as rapid deforestation and agricultural expansion—have resulted in humans and animals coming into more frequent and intense contact with one another, increasing opportunities for what is known as "zoonotic disease spillover." In the past few years alone, numerous disease outbreaks have had suspected or confirmed zoonotic origin, including mpox (formerly known as monkeypox), Ebola virus disease, dengue fever, and COVID-19. Experts also recognize the need to prepare for another possible Disease X, a term used to describe a currently unknown pathogen with pandemic potential. To direct resources toward the most high-consequence pathogens, it is paramount that leaders have an accurate concept of pandemic risk—for individual viruses as well as viral families. Several institutions are developing disease rankings at national and global levels, including the Priority Zoonotic Diseases Lists facilitated by the U.S. Centers for Disease Control and Prevention and the Research and Development (R&D) Blueprint created by the World Health Organization. The original SpillOver risk ranking framework (SpillOver 1.0), an open-source webtool launched by researchers at the University of California, Davis One Health Institute, estimated the relative spillover potential of wildlife-origin viruses to humans based on a series of host, viral, and environmental risk factors determined via expert opinion and scientific evidence. Its next iteration, SpillOvers 2.0, has rebranded to better describe the diversity and frequency of virus spillovers to people. The new platform uses a One Health approach, which recognizes the interdependence of human, animal, and environmental health. It will expand to include domestic animal and vector-borne viruses and assess pandemic risk rather than just spillover risk for wildlife viruses.

|
Scooped by
nrip
July 24, 2021 9:24 AM
|
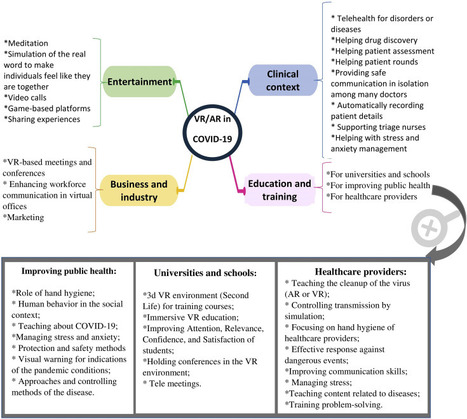
The pandemics of major infectious diseases often cause public health, economic, and social problems. Virtual reality (VR) and augmented reality (AR), as two novel technologies, have been used in many fields for emergency management of disasters. The objective of this paper was to review VR and AR applications in the emergency management of infectious outbreaks with an emphasis on the COVID-19 outbreak. It appears that VR and AR technologies can play a positive role during infectious disease outbreaks. VR and AR have been widely used in the prevention and response phases of emergency management during infectious disease pandemics, such as SARS and Ebola pandemics, especially for educating and training purposes for the public. During the COVID-19 outbreak, these technologies have the potential to be used in various fields, including 1) clinical context (e.g., telehealth, drug discovery, patient assessment, mental health management), 2) entertainment (e.g., video call, meditation, gaming), 3) business and industry (e.g., holding meetings and conferences, marketing), and 4) education (e.g., in schools and universities, for healthcare providers, and VR-based content for improving public health). These technologies can be used in the above-mentioned fields by providing their different features for facilitating the challenges of COVID-19. However, to respond to COVID-19, all applications of VR and AR should be considered as a supportive approach alongside other information technologies. We believe that VR and AR have a substantial potential to impact the emergency management of COVID-19 or any infectious disease pandemics; however, these potentials need to be studied in a more robust manner. read the paper ta https://www.sciencedirect.com/science/article/pii/S2352914821000691

|
Scooped by
nrip
July 16, 2021 4:33 PM
|
The national severe acute respiratory illness (SARI) surveillance system in Yemen was established in 2010 to monitor SARI occurrence in humans and provide a foundation for detecting SARI outbreaks. This study aimed to examine the level of usefulness and the performance of the SARI surveillance system in Yemen.
Methods: The updated Centers for Disease Control and Prevention guidelines were used for the purposes of our evaluation. Related documents and reports were reviewed. Data were collected from 4 central-level managers and stakeholders and from 10 focal points at 4 sentinel sites by using a semistructured questionnaire.
For each attribute, percent scores were calculated and ranked as follows: very poor (≤20%), poor (20%-40%), average (40%-60%), good (60%-80%), and excellent (>80%). The surveillance systems at sentinel sites are tools for the early detection of disease, the monitoring of trends in the burden of diseases, and the generation of recommendations for the prevention and control of diseases. The evaluation of surveillance systems helps decision makers to set priorities for future planning, resource allocation, and future interventions for preventing the spread of diseases.
Results: As rated by the evaluators, the SARI surveillance system achieved its objectives.
The system’s flexibility (percent score: 86%) and acceptability (percent score: 82%) were rated as “excellent,” and simplicity (percent score: 74%) and stability (percent score: 75%) were rated as “good.” The percent score for timeliness was 23% in 2018, which indicated poor timeliness. The overall data quality percent score of the SARI system was 98.5%. Despite its many strengths, the SARI system has some weaknesses. For example, it depends on irregular external financial support.
The SARI surveillance system was useful in estimating morbidity and mortality, monitoring the trends of the disease, and promoting research for informing prevention and control measures.
The overall performance of the SARI surveillance system was good. We recommend expanding the system by promoting private health facilities’ (eg, private hospitals and private health centers) engagement in SARI surveillance, establishing an electronic database at central and peripheral sites, and providing the National Central Public Health Laboratory with the reagents needed for disease confirmation.

|
Scooped by
nrip
July 15, 2021 2:15 AM
|
Agencies such as the Centers for Disease Control and Prevention (CDC) currently release influenza-like illness incidence data, along with descriptive summaries of simple spatio-temporal patterns and trends. However, public health researchers, government agencies, as well as the general public, are often interested in deeper patterns and insights into how the disease is spreading, with additional context. Analysis by domain experts is needed for deriving such insights from incidence data.
Objective: Our goal was to develop an automated approach for finding interesting spatio-temporal patterns in the spread of a disease over a large region, such as regions which have specific characteristics (eg, high incidence in a particular week, those which showed a sudden change in incidence) or regions which have significantly different incidence compared to earlier seasons.
Methods: We developed techniques from the area of transactional data mining for characterizing and finding interesting spatio-temporal patterns in disease spread in an automated manner. A key part of our approach involved using the principle of minimum description length for representing a given target set in terms of combinations of attributes (referred to as clauses); we considered both positive and negative clauses, relaxed descriptions which approximately represent the set, and used integer programming to find such descriptions. Finally, we designed an automated approach, which examines a large space of sets corresponding to different spatio-temporal patterns, and ranks them based on the ratio of their size to their description length (referred to as the compression ratio).
Results: We applied our methods using minimum description length to find spatio-temporal patterns in the spread of seasonal influenza in the United States using state level influenza-like illness activity indicator data from the CDC. We observed that the compression ratios were over 2.5 for 50% of the chosen sets, when approximate descriptions and negative clauses were allowed. Sets with high compression ratios (eg, over 2.5) corresponded to interesting patterns in the spatio-temporal dynamics of influenza-like illness. Our approach also outperformed description by solution in terms of the compression ratio.
Conclusions: Our approach, which is an unsupervised machine learning method, can provide new insights into patterns and trends in the disease spread in an automated manner. Our results show that the description complexity is an effective approach for characterizing sets of interest, which can be easily extended to other diseases and regions beyond influenza in the US. Our approach can also be easily adapted for automated generation of narratives. read more at https://publichealth.jmir.org/2020/3/e12842

|
Rescooped by
nrip
from healthcare technology
June 17, 2021 12:25 AM
|
We discuss the concept of a participatory digital contact notification approach to assist tracing of contacts who are exposed to confirmed cases of coronavirus disease (COVID-19); The core functionality of our concept is to provide a usable, labor-saving tool for contact tracing by confirmed cases themselves the approach is simple and affordable for countries with limited access to health care resources and advanced technology. The proposed tool serves as a supplemental contract tracing approach to counteract the shortage of health care staff while providing privacy protection for both cases and contacts. - This tool can be deployed on the internet or as a plugin for a smartphone app.
- Confirmed cases with COVID-19 can use this tool to provide contact information (either email addresses or mobile phone numbers) of close contacts.
- The system will then automatically send a message to the contacts informing them of their contact status, what this status means, the actions that should follow (eg, self-quarantine, respiratory hygiene/cough etiquette), and advice for receiving early care if they develop symptoms.
- The name of the sender of the notification message by email or mobile phone can be anonymous or not.
- The message received by the contact contains no disease information but contains a security code for the contact to log on the platform to retrieve the information.
Conclusion The successful application of this tool relies heavily on public social responsibility and credibility, and it remains to be seen if the public would adopt such a tool and what mechanisms are required to prevent misuse. This is a simple tool that does not require complicated computer techniques despite strict user privacy protection design with respect to countries and regions. Additionally, this tool can help avoid coercive surveillance, facilitate the allocation of health resources, and prioritize clinical service for patients with COVID-19. Information obtained from the platform can also increase our understanding of the epidemiology of COVID-19. read this concept paper at https://mhealth.jmir.org/2020/6/e20369

|
Scooped by
nrip
June 15, 2021 6:44 AM
|
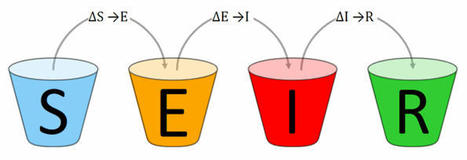
Pandemic SEIR and SEIRV modelling software and infrastructure for the Corona SARS-COV-2 COVID-19 disease with data from Johns-Hopkins-University CSSE, Robert Koch-Institute and vaccination data from Our World In Data. The SARS-COV-2 pandemic has been affecting our lives for months. The effectiveness of measures against the pandemic can be tested and predicted by using epidemiological models. The Corona SEIR Workbench uses a SEIR model and combines a graphical output of the results with a simple parameter input for the model. Modelled data can be compared country by country with the SARS-COV-2 infection data of the Johns Hopkins University. Additionally, the R₀ values of the Robert Koch Institute can be displayed for Germany. Vaccination data is used from Our World In Data.

|
Scooped by
nrip
June 1, 2021 4:24 PM
|
Despite recent achievements in vaccines, antiviral drugs, and medical infrastructure, the emergence of COVID-19 has posed a serious threat to humans worldwide. Most countries are well connected on a global scale, making it nearly impossible to implement perfect and prompt mitigation strategies for infectious disease outbreaks. In particular, due to the explosive growth of international travel, the complex network of human mobility enabled the rapid spread of COVID-19 globally.
Objective:
South Korea was one of the earliest countries to be affected by COVID-19. In the absence of vaccines and treatments, South Korea has implemented and maintained stringent interventions, such as large-scale epidemiological investigations, rapid diagnosis, social distancing, and prompt clinical classification of severely ill patients with appropriate medical measures. In particular, South Korea has implemented effective airport screenings and quarantine measures. In this study, we aimed to assess the country-specific importation risk of COVID-19 and investigate its impact on the local transmission of COVID-19. Methods: The country-specific importation risk of COVID-19 in South Korea was assessed. We investigated the relationships between country-specific imported cases, passenger numbers, and the severity of country-specific COVID-19 prevalence from January to October 2020. We assessed the country-specific risk by incorporating country-specific information. A renewal mathematical model was employed, considering both imported and local cases of COVID-19 in South Korea. Furthermore, we estimated the basic and effective reproduction numbers.
Results: The risk of importation from China was highest between January and February 2020, while that from North America (the United States and Canada) was high from April to October 2020. The R0 was estimated at 1.87 (95% CI 1.47-2.34), using the rate of α=0.07 for secondary transmission caused by imported cases. The Rt was estimated in South Korea and in both Seoul and Gyeonggi.
Conclusions: A statistical model accounting for imported and locally transmitted cases was employed to estimate R0 and Rt.
Our results indicated that the prompt implementation of airport screening measures (contact tracing with case isolation and quarantine) successfully reduced local transmission caused by imported cases despite passengers arriving from high-risk countries throughout the year. Moreover, various mitigation interventions, including social distancing and travel restrictions within South Korea, have been effectively implemented to reduce the spread of local cases in South Korea. read the paper at https://publichealth.jmir.org/2021/6/e26784/

|
Scooped by
nrip
May 15, 2021 12:52 PM
|
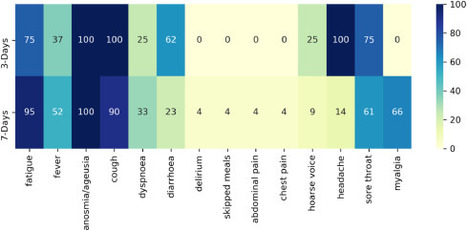
In a clinical trial, diagnostic testing of suspected cases e.g RT-PCR for severe acute respiratory SARS-CoV-2, can be triggered by the presence of any COVID-19 associated symptom.
A household survey in the United Kingdom (UK) showed that fever, cough, anosmia, and ageusia were present on the day of testing in only 60% of symptomatic, RT-PCR positive individuals, implying that other less specific signs/symptoms associated with COVID-19 occur in a substantial number of patients.
The signs/symptoms associated with COVID-19 are extensive and overlap with those of other common viral infections.
Diagnostic work-up following any COVID-19 associated symptom will lead to extensive testing, potentially overwhelming laboratory capacity whilst primarily yielding negative results. We aimed to identify optimal symptom combinations to capture most cases using fewer tests with implications for COVID-19 vaccine developers across different resource settings and public health.
Methods
UK and US users of the COVID-19 Symptom Study app who reported new-onset symptoms and an RT-PCR test within seven days of symptom onset were included. Sensitivity, specificity, and number of RT-PCR tests needed to identify one case (test per case [TPC]) were calculated for different symptom combinations.
A multi-objective evolutionary algorithm was applied to generate combinations with optimal trade-offs between sensitivity and specificity.
Interpretation
We confirmed the significance of COVID-19 specific symptoms for triggering RT-PCR and identified additional symptom combinations with optimal trade-offs between sensitivity and specificity that maximize case capture given different resource settings.
read this paper at https://www.journalofinfection.com/article/S0163-4453(21)00079-7/fulltext

|
Scooped by
nrip
May 12, 2021 8:55 AM
|
Given the potential for the evolution of SARS-CoV-2 variants that render vaccines less effective (vaccine escape), assisted by waning immunity following vaccination, how can COVID-19 exit strategies be planned while limiting the vaccine escape risk? A key component of any plausible strategy towards the permanent removal of non-pharmaceutical interventions (NPIs) is ensuring low case numbers in the short to medium term using NPIs and vaccination. Read this paper which discusses this mathematically https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00202-4/fulltext
|

|
Scooped by
nrip
September 15, 2025 11:39 PM
|
The integration of viral genomic data into public health surveillance has revolutionized our ability to track and forecast infectious disease dynamics. This review addresses two critical aspects of infectious disease forecasting and monitoring: the methodological workflow for epidemic forecasting and the transformative role of molecular surveillance. Further it : -
Provides comprehensive, iterative workflow for reliably fitting and forecasting epidemic models, emphasizing key stages such as data integration, model refinement, and thorough parameter identifiability assessments. - •
Underscores the importance of both structural and practical identifiability analyses to ensure robust parameter estimation and uncertainty quantification, thereby improving the reliability of epidemic forecasts. - •
Highlights the role of molecular surveillance, particularly genomic epidemiology, as a fundamental tool for enabling real-time tracking of pathogen evolution and spread with unparalleled precision and accuracy. - •
Addresses computational complexities associated with integrating genomic data into traditional epidemiological models and proposes strategies to overcome these challenges to optimize model performance. read the entire piece at https://www.sciencedirect.com/science/article/pii/S1571064524001350

|
Scooped by
nrip
January 16, 2025 12:17 AM
|
An urgent need of implementing national surveillance systems for timely detection and reporting of emerging antimicrobial resistance (AMR) was recently advocated by the World Health Organization (WHO). However, public information on existing national early warning systems (EWSs) is often incomplete, and a comprehensive overview on this topic is currently lacking. Objective of this study: To map the availability of EWSs for emerging AMR in high-income countries and describe their main characteristics. Methods: A systematic review was performed on bibliographic databases, and a targeted search was conducted on national websites. Any article, report or webpage describing national EWSs in high-income countries was eligible for inclusion. EWSs were identified considering the emerging AMR reporting WHO framework. Results: We identified seven national EWSs in 72 high-income countries: two (Australia, Japan) in the East Asia and Pacific Region, three (France, Sweden, United Kingdom) in Europe and Central Asia, and two (United States, Canada) in North America. The systems were established quite recently; in most cases they covered both community and hospital settings, but their main characteristics varied widely across countries in terms of organization and microorganisms under surveillance, with also different definitions of emerging AMR and alert functioning. A formal system assessment was available only in Australia. Conclusions: A broader implementation and investment of national surveillance systems that allow early detection of emerging AMR is still needed to establish EWSs in countries and regions lacking such capabilities. More standardized data collection and reporting are also advisable to improve cooperation on a global scale. Further research is required to provide an in-depth analysis of EWSs, as this study is limited to publicly available data on HICs.

|
Scooped by
nrip
October 7, 2024 1:17 PM
|
Research that spans clinical specialties and research disciplines beyond health and healthcare is a priority for planning equitable responses to manage future health shocks, argue Amitava Banerjee and colleagues Health shocks are “high consequence events that have a major disruptive effect on society,”1 with health, social, economic, and psychological effects, and are not limited to pandemics. Whether responding to shocks related to antimicrobial resistance, climate change, or conflict, siloed research cannot deliver the science needed quickly enough at the required scale. Recommendations for multidisciplinary, interdisciplinary, and transdisciplinary research for future shocks Multidisciplinary research Science—Collaborate and integrate across health data related disciplines, organisations, and health data for research. Governments, regulators, and funders should facilitate national level, linked electronic health record research across disciplines for future shocks. Evidence—Avoid unnecessary communicable versus non-communicable disease dichotomies. Guideline committees and professional societies relating to shocks should consider how both communicable and non-communicable diseases can and should be incorporated. Care—Unify methodology and approach across disciplines to tackle indirect effects of shocks. Governments should ensure representation in advisory committees by all stakeholders, including medical specialties, research disciplines, and expertise relating to different shocks to have strategies to reduce and tackle indirect effects in shock preparedness. Interdisciplinary research Science—Investigate long term impact of shocks on acute and long term conditions and their care. Funders should prioritise specific research calls in long term effects for shock preparedness and also during acute phases of shocks. Evidence—Prioritise patient and public involvement in all areas of research and guideline development in relation to shocks. Mandates should involve patients and public, by academics in interdisciplinary research, and by governments and professional societies in guidelines. Care—Recognise that population subgroups may respond differently to specific interventions, developing culturally sensitive strategies where necessary. Governments and funders should prioritise national data collection before, during, and after shocks relating to protected characteristics, including age, ethnicity, and socioeconomic status. Transdisciplinary research Science—Tackle social, cultural, and economic determinants of health by developing an urgent, well resourced and prioritised transdisciplinary research strategy, including focus on long term societal impact of shocks. Governments should set up a commission, involving stakeholders (including research funders and organisations) to develop a transdisciplinary research strategy for impact of shocks. Evidence—Use large scale, representative, linked electronic health record data, managed and led by multidisciplinary teams, including public representatives. Governments and research funders should facilitate and resource linked data research across health and non-health disciplines in relation to shocks. Care—Ensure wide angle views of health inequalities and patient/public involvement to produce effective and acceptable strategies. Funders should have specific funding calls for health inequalities during shocks to foster representative, inclusive patient and public involvement. read more at https://www.bmj.com/content/387/bmj-2023-078445.short?rss=1

|
Scooped by
nrip
September 19, 2024 11:18 PM
|

|
Rescooped by
nrip
from Healthcare in India
July 29, 2021 10:31 AM
|
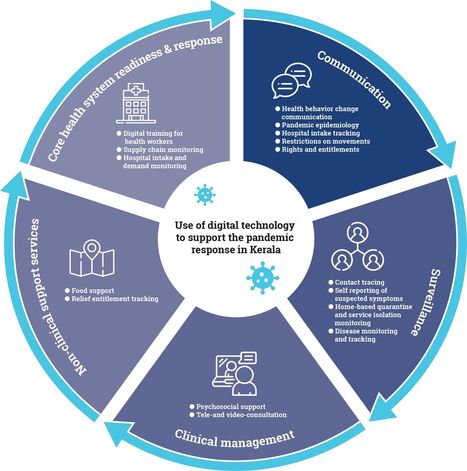
Digital tools are increasingly being applied to support the response to the ongoing COVID-19 pandemic in India and elsewhere globally. This article draws from global frameworks to explore the use of digital tools in the state of Kerala across the domains of communication, surveillance, clinical management, non-clinical support, and core health system readiness and response. Kerala is considered India’s first digital state, with the highest percentage of households with computers (24%) and the internet (51%) in India, 95% mobile phone penetration, 62% smartphone penetration and 75% digital literacy. Kerala has long been a model for the early adoption of digital technology for education and health. As part of the pandemic response, technology has been used across private and public sectors, including law enforcement, health, information technology and education. Efforts have sought to ensure timely access to health information, facilitate access to entitlements, monitor those under quarantine and track contacts, and provide healthcare services though telemedicine. Kerala’s COVID-19 pandemic response showcases the diverse potential of digital technology, the importance of building on a strong health system foundation, the value of collaboration, and the ongoing challenges of data privacy and equity in digital access. Summary -
The COVID-19 pandemic’s unprecedented global spread and impact has accelerated interest in digital innovation. -
Kerala’s experience showcases the diverse and innovative ways that digital tools can build on a strong underlying health system to support pandemic response across the domains of communication, surveillance, clinical management, non-clinical support and core health system readiness. -
Digital tools in Kerala were able to proliferate rapidly and help meet diverse citizen needs due to high levels of collaboration and intersectoral response that brought together different levels of government and multiple state departments, engaged the private sector, and harnessed the energy of civil society organisations and community volunteers. -
Digital technology has great potential to strengthen public health measures during pandemics, including to rapidly link citizens to food and mental health support. -
Adequate oversight and community participation remains essential to safeguard citizen privacy and ensure equity. read the open access paper at https://gh.bmj.com/content/6/Suppl_5/e005355

|
Scooped by
nrip
July 19, 2021 10:52 PM
|
A team of researchers analyzed the genomes of more than 2,500 modern humans from 26 worldwide populations, to better understand how humans have adapted to historical coronavirus outbreaks. The team used computational methods to uncover genetic traces of adaptation to coronaviruses, the family of viruses responsible for three major outbreaks in the last 20 years, including the ongoing COVID-19 pandemic. Traces of the outbreak are evident in the genetic makeup of people from that area, they’ve found. A coronavirus epidemic broke out in the East Asia region more than 20,000 years ago, as per their findings. The discovery of a coronavirus outbreak from 20,000 years ago is "like finding fossilized dinosaur footprints instead of finding fossilized bones directly. The work shows that over the course of the epidemic, selection favored certain variants of human genes involved in the virus-cell interactions that could have led to a less severe disease. Studying the “tracks” left by ancient viruses can help researchers better understand how the genomes of different human populations adapted to viruses that have emerged as important drivers of human evolution. The study’s authors say their research could help identify viruses that have caused epidemics in the distant past and may do so in the future. Studies like theirs help researchers compile a list of potentially dangerous viruses and then develop diagnostics, vaccines, and drugs for the event of their return. read the paper at https://www.cell.com/current-biology/fulltext/S0960-9822(21)00794-6 more at https://www.futurity.org/coronavirus-epidemic-viruses-2597742/

|
Scooped by
nrip
July 15, 2021 3:29 PM
|
This study aims to describe the compounding factors in a complex emergency, which exacerbate a cholera epidemic among vulnerable populations due to supply chain disruptions. Basic needs such as food, medicine, water, sanitation and hygiene commodities are critical to reduce the incidence rate of cholera and control the spread of infection. Conflicts cause damage to infrastructure, displace vulnerable populations and restrict the flow of goods from both commercial and humanitarian organizations. This study assesses the underlying internal and external factors that either aggravate or mitigate the risk of a cholera outbreak in such settings, using Yemen as a case study. Findings Compounding factors that influenced the cholera outbreak in Yemen are visualized in a causal loop diagram, which can improve the understanding of relationships where numerous uncertainties exist. A strong link exists between humanitarian response and the level of infrastructure development in a country. Supply chains are affected by constraints deriving from the Yemeni conflict, further inhibiting the use of infrastructure, which limits access to basic goods and services. Aligning long-term development objectives with short-term humanitarian response efforts can create more flexible modes of assistance to prevent and control future outbreaks. Practical implications This study presents a systematic view of dynamic factors existing in complex emergencies that have cause-and-effect relationships. Several models of cholera outbreaks have been used in previous studies, primarily focusing on the modes and mechanisms of transmission throughout a population. However, such models typically do not include other internal and external factors that influence the population and context at the site of an outbreak. This model incorporates those factors from a logistics perspective to address the distribution of in-kind goods and cash and voucher assistance. Social implications This study has been aligned with six of the United Nations Sustainable Development Goals (SDGs), using their associated targets in the model as variables that influence the cholera incidence rate. Recognizing that the SDGs are interlinked, as are the dynamic factors in complex humanitarian emergencies, the authors have chosen to take an interdisciplinary approach to consider social, economic and environmental factors that may be impacted by this research. read more at https://www.emerald.com/insight/content/doi/10.1108/JHLSCM-07-2020-0063/full/html

|
Rescooped by
nrip
from healthcare technology
June 17, 2021 12:26 AM
|
The COVID-19 pandemic is the greatest public health crisis of the last 100 years. Countries have responded with various levels of lockdown to save lives and stop health systems from being overwhelmed. At the same time, lockdowns entail large socioeconomic costs. One exit strategy under consideration is a mobile phone app that traces the close contacts of those infected with COVID-19. Recent research has demonstrated the theoretical effectiveness of this solution in different disease settings. However, concerns have been raised about such apps because of the potential privacy implications. This could limit the acceptability of app-based contact tracing in the general population. As the effectiveness of this approach increases strongly with app uptake, it is crucial to understand public support for this intervention. Objective: The objective of this study is to investigate the user acceptability of a contact-tracing app in five countries hit by the pandemic.
Methods: We conducted a largescale, multicountry study (N=5995) to measure public support for the digital contact tracing of COVID-19 infections.
We ran anonymous online surveys in France, Germany, Italy, the United Kingdom, and the United States and measured intentions to use a contact-tracing app across different installation regimes (voluntary installation vs automatic installation by mobile phone providers) and studied how these intentions vary across individuals and countries.
Results: We found strong support for the app under both regimes, in all countries, across all subgroups of the population, and irrespective of regional-level COVID-19 mortality rates.
We investigated the main factors that may hinder or facilitate uptake and found that concerns about cybersecurity and privacy, together with a lack of trust in the government, are the main barriers to adoption.
Conclusions:
Epidemiological evidence shows that app-based contact tracing can suppress the spread of COVID-19 if a high enough proportion of the population uses the app and that it can still reduce the number of infections if uptake is moderate. Our findings show that the willingness to install the app is very high. The available evidence suggests that app-based contact tracing may be a viable approach to control the diffusion of COVID-19. read the study at https://mhealth.jmir.org/2020/8/e19857

|
Rescooped by
nrip
from healthcare technology
June 17, 2021 12:23 AM
|
Patient travel history can be crucial in evaluating evolving infectious disease events. Such information can be challenging to acquire in electronic health records, as it is often available only in unstructured text.
Objective: This study aims to assess the feasibility of annotating and automatically extracting travel history mentions from unstructured clinical documents in the Department of Veterans Affairs across disparate health care facilities and among millions of patients. Information about travel exposure augments existing surveillance applications for increased preparedness in responding quickly to public health threats.
Methods: Clinical documents related to arboviral disease were annotated following selection using a semiautomated bootstrapping process. Using annotated instances as training data, models were developed to extract from unstructured clinical text any mention of affirmed travel locations outside of the continental United States. Automated text processing models were evaluated, involving machine learning and neural language models for extraction accuracy.
Results: Among 4584 annotated instances, 2659 (58%) contained an affirmed mention of travel history, while 347 (7.6%) were negated. Interannotator agreement resulted in a document-level Cohen kappa of 0.776. Automated text processing accuracy (F1 85.6, 95% CI 82.5-87.9) and computational burden were acceptable such that the system can provide a rapid screen for public health events.
Conclusions: Automated extraction of patient travel history from clinical documents is feasible for enhanced passive surveillance public health systems.
Without such a system, it would usually be necessary to manually review charts to identify recent travel or lack of travel, use an electronic health record that enforces travel history documentation, or ignore this potential source of information altogether. The development of this tool was initially motivated by emergent arboviral diseases. More recently, this system was used in the early phases of response to COVID-19 in the United States, although its utility was limited to a relatively brief window due to the rapid domestic spread of the virus. Such systems may aid future efforts to prevent and contain the spread of infectious diseases. read the study at https://publichealth.jmir.org/2021/3/e26719

|
Scooped by
nrip
June 10, 2021 2:23 AM
|
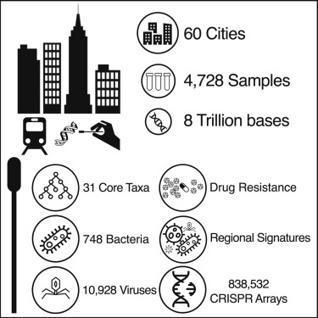
Here is a global atlas of 4,728 metagenomic samples from mass-transit systems in 60 cities over 3 years, representing the first systematic, worldwide catalog of the urban microbial ecosystem. This atlas provides an annotated, geospatial profile of microbial strains, functional characteristics, antimicrobial resistance (AMR) markers, and genetic elements, including 10,928 viruses, 1,302 bacteria, 2 archaea, and 838,532 CRISPR arrays not found in reference databases. Authors identified 4,246 known species of urban microorganisms and a consistent set of 31 species found in 97% of samples that were distinct from human commensal organisms. Profiles of AMR genes varied widely in type and density across cities. Cities showed distinct microbial taxonomic signatures that were driven by climate and geographic differences. These results constitute a high-resolution global metagenomic atlas that enables discovery of organisms and genes, highlights potential public health and forensic applications, and provides a culture-independent view of AMR burden in cities.

|
Scooped by
nrip
May 16, 2021 6:38 PM
|
Background: In March 2020, South Africa implemented strict nonpharmaceutical interventions (NPIs) to contain the spread of COVID-19. Over the subsequent 5 months, NPI policies were eased in stages according to a national strategy. COVID-19 spread throughout the country heterogeneously; the case numbers peaked from July to August. A second COVID-19 wave began in late 2020. Data on the impact of NPI policies on social and economic well-being and access to health care are limited.
Objective: We aimed to determine how rural residents in three South African provinces changed their behaviors during the first COVID-19 epidemic wave.
Methods: The South African Population Research Infrastructure Network nodes in the Mpumalanga (Agincourt), KwaZulu-Natal, (Africa Health Research Institute) and Limpopo (Dikgale-Mamabolo-Mothiba) provinces conducted up to 14 rounds of longitudinal telephone surveys among randomly sampled households from rural and periurban surveillance populations every 2-3 weeks. Interviews included questions on the following topics: COVID-19–related knowledge and behaviors, the health and economic impacts of NPIs, and mental health. We analyzed how responses varied based on NPI stringency and household sociodemographics.
Conclusions: South Africans complied with stringent, COVID-19–related NPIs despite the threat of substantial social, economic, and health repercussions. Government-supported social welfare programs appeared to buffer interruptions in income and health care access during local outbreaks. Epidemic control policies must be balanced against the broader well-being of people in resource-limited settings and designed with parallel support systems when such policies threaten peoples’ income and access to basic services.

|
Scooped by
nrip
May 13, 2021 12:35 PM
|
The major medical and social challenge of the 21st century is COVID-19, caused by the novel coronavirus SARS-CoV-2. Critical issues include the rate at which the coronavirus spreads and the effect of quarantine measures and population vaccination on this rate. Knowledge of the laws of the spread of COVID-19 will enable assessment of the effectiveness and reasonableness of the quarantine measures used, as well as determination of the necessary level of vaccination needed to overcome this crisis. Objective: This study aims to establish the laws of the spread of COVID-19 and to use them to develop a mathematical model to predict changes in the number of active cases over time, possible human losses, and the rate of recovery of patients, to make informed decisions about the number of necessary beds in hospitals, the introduction and type of quarantine measures, and the required threshold of vaccination of the population. Methods: This study analyzed the onset of COVID-19 spread in countries such as China, Italy, Spain, the United States, the United Kingdom, Japan, France, and Germany based on publicly available statistical data. The change in the number of COVID-19 cases, deaths, and recovered persons over time was examined, considering the possible introduction of quarantine measures and isolation of infected people in these countries. Based on the data, the virus transmissibility and the average duration of the disease at different stages were evaluated, and a model based on the principle of recursion was developed. Its key features are the separation of active (nonisolated) infected persons into a distinct category and the prediction of their number based on the average duration of the disease in the inactive phase and the concentration of these persons in the population in the preceding days. Results: Specific values for SARS-CoV-2 transmissibility and COVID-19 duration were estimated for different countries. In China, the viral transmissibility was 3.12 before quarantine measures were implemented and 0.36 after these measures were lifted. For the other countries, the viral transmissibility was 2.28-2.76 initially, and it then decreased to 0.87-1.29 as a result of quarantine measures. Therefore, it can be expected that the spread of SARS-CoV-2 will be suppressed if 56%-64% of the total population becomes vaccinated or survives COVID-19. Conclusions: The quarantine measures adopted in most countries are too weak compared to those previously used in China. Therefore, it is not expected that the spread of COVID-19 will stop and the disease will cease to exist naturally or owing to quarantine measures. Active vaccination of the population is needed to prevent the spread of COVID-19. Furthermore, the required specific percentage of vaccinated individuals depends on the magnitude of viral transmissibility, which can be evaluated using the proposed model and statistical data for the country of interest. read the entire paper at https://publichealth.jmir.org/2021/4/e21468/
|


 Your new post is loading...
Your new post is loading...