Your new post is loading...
 Your new post is loading...

|
Scooped by
I2BC Paris-Saclay
May 7, 4:30 AM
|
New training session on FIB-SEM applications to biology at ambient temperature

|
Scooped by
I2BC Paris-Saclay
April 2, 8:28 AM
|
The I2BC crystallization platform is evolving into a crystallography platform, offering new services
The crystallization platform has evolved into a crystallography platform and now offers new services (including protein structure determination and/or analysis) as well as scientific support for clients in structural biology projects. Protein crystallography is the main approach for characterizing the 3D structure of proteins associated with drug candidates and for determining the molecular basis of complexes between soluble or membrane proteins and their partners (DNA, RNA, other proteins, peptides, lipids, cofactors, various ligands including metals). Initially, the crystallization platform was dedicated to automated screening, analysis, and optimization of crystallization conditions for soluble and membrane macromolecules. Now, the crystallography platform provides extended services, including protein structure determination and/or analysis. The successive steps, starting from a purified protein sample of interest, include crystal formation, diffraction image collection at the Synchrotron Soleil, electron density map calculation, atomic model refinement of the protein or complex of interest, and structural analysis. Emphasis is placed on obtaining the most accurate models for the most successful structural analysis. Additionally, an AlphaFold service, including predicted model analysis, is available, relying on services established at I2BC by the BIOI2 integrative bioinformatics platform. Routine upstream steps before obtaining a protein crystal include obtaining an optimized synthetic gene for expression in prokaryotic hosts, expression, and purification of the recombinant protein of interest. The platform can advise clients on these various steps, and collaboration or service provision can be considered depending on project complexity. The crystallography platform is open to all scientists, from beginners to experienced crystallographers. It is important to note that the quality of the protein sample is a crucial criterion. The sample must be pure and verified on a denaturing acrylamide gel before crystallization. Do not hesitate to contact us. Contact: cristallographie@i2bc.paris-saclay.fr
More information: https://www.i2bc.paris-saclay.fr/structural-biology/crystallography/

|
Scooped by
I2BC Paris-Saclay
January 16, 8:23 AM
|
Newcomer at the BIOI2 bioinformatics platform
Baptiste Roelens joined the BIOI2 platform last month, bringing expertise in machine learning for biological data analysis. His contributions will expand the platform's service offerings and support the I2BC imaging community. The integrative BIOinformatics platform (BIOI2) welcomes Baptiste Roelens as a new engineer. Baptiste's background combines theoretical computer science and life sciences, with a foundation from Ecole Polytechnique. He further honed his skills at the Institut Curie, where he earned his PhD. His doctoral research focused on developing quantitative approaches to analyze the role of the histone chaperone CAF-1 in maintaining higher-order chromatin structure during meiotic prophase in the fruit fly. Continuing his research journey, Baptiste joined Stanford University to develop bioimage analysis pipelines, further exploring the mechanisms regulating chromosome organization and cell-cycle progression in C. elegans germline. Upon returning to France, he contributed to the biotech company Viroxis, developing cutting-edge deep-learning approaches for protein design. With experience spanning both wet and dry labs, Baptiste will be a great asset to the BIOI2 platform and is ideally placed to collaborate with the I2BC’s imaging community, in particular the Imagerie-Gif platform (https://www.i2bc.paris-saclay.fr/bioimaging/), to develop pipelines that streamline the processing and analysis of complex imaging data. BIOI2 provides access to bioinformatics resources and activities developed and of use at the I2BC, organises focused training session on bioinformatics tools, and is an official contributing plateform of the nationwide French Institute of Bioinformatics (IFB). More information: https://bioi2.i2bc.paris-saclay.fr Contact: contact-bioi2@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
July 26, 2024 8:24 AM
|
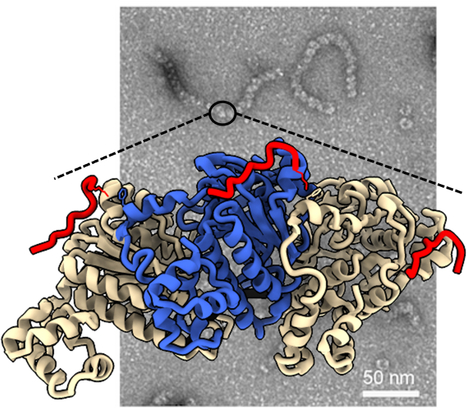
BRCA2 stabilizes DMC1 nucleoprotein filaments in meiosis
To repair programmed double-strand breaks in meiosis, the DNA repair protein BRCA2 binds to the recombinase DMC1 either monomeric or assembled on single-stranded DNA through two different interfaces, and stabilizes DMC1 nucleoprotein filaments. The BReast CAncer type 2 susceptibility protein (BRCA2), a tumor suppressor mutated in breast, ovarian and prostate cancers, plays a major role in the repair of DNA double-strand breaks by homologous recombination, both in somatic cells and during meiosis. BRCA2 interacts with the ubiquitous recombinase RAD51 , as well as the meiotic recombinase DMC1, and facilitates their loading at double-strand break sites. BRCA2 interacts with these recombinases via FxxA and FxPP motifs (called A and P motifs, respectively). In a study published recently in the journal Nucleic Acids Research, scientists from the INTGEN team at the I2BC (Université Paris Saclay, CEA, CNRS, Gif-sur-Yvette) and the PROXIMA-1 beamline at the SOLEIL synchrotron (Université Paris Saclay) solved the crystal structure of the complex between a BRCA2 fragment containing a P motif (PhePP) and the DMC1 protein. Together with the team of A. Zelensky and R. Kanaar (Erasmus Medical Center, Rotterdam), they showed that A and P motifs bind to distinct sites on the ATPase domain of recombinases. The P motif interacts with a site that is accessible in the octamers of DMC1 and the nucleoprotein filaments formed by DMC1 loaded onto single-stranded DNA. Furthermore, in collaboration with scientists from the Institut Gustave Roussy (Université Paris Saclay), they revealed that this interaction involves two adjacent DMC1 protomers, thereby increasing the stability of the nucleoprotein filaments. These results help to explain why the region encoded by exons 12 to 14 of the BRCA2 gene (the PhePP motif being encoded by exon 14) is essential during meiotic homologous recombination in mice (work by the teams of A. Zelensky and W. Baarends, Erasmus Medical Center, Rotterdam, Netherlands). More information: https://doi.org/10.1093/nar/gkae452 Contact: Simona MIRON <simona.miron@i2bc.paris-saclay.fr> & Sophie ZINN <sophie.zinn@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
January 30, 2024 10:07 AM
|
Nikon presents the AX-NSPARC at Imagerie-Gif - From January 30 to February 9 -
Nikon is presenting its latest confocal microscope to the Paris-Saclay community, equipped with a new detector for very high-resolution images (XY : 100 nm). From January 30 to February 9, 8 days of demonstrations on the Gif Imaging platform will enable you to test the microscope with your own samples . Don't hesitate to come and take advantage of this opportunity. More information: here

|
Scooped by
I2BC Paris-Saclay
December 1, 2023 9:15 AM
|
New: NGS small RNA library preparation training
The I2BC NGS facility organizes a new training on the preparation of small RNA libraries with technical tips and tricks to conduct successful small RNA NGS experiments. MicroRNAs (miRNAs) and other types of small RNA (sRNA) molecules play important roles in the cell and their dysregulation has been linked to various diseases. Indeed, circulating miRNAs are found in various human body fluids and are of interest as potential new non-invasive biomarkers. Next-generation sequencing (NGS) allows a very sensitive and quantitative detection of sRNA expression profiles. However, NGS library preparation from sRNAs is technically relatively challenging as compared to NGS protocols for other applications. Classical sRNA library preparation methods often introduce serious biases leading to sequencing data that do not represent the original sRNA expression profiles. The new training organized by the I2BC NGS facility focusses on technical aspects of NGS library preparation from different types of sRNAs. These include natural sRNAs such as miRNAs, and artificial sRNAs such as ribosome-protected fragments (Ribo-seq) or co-immunoprecipitated sRNA fragments (e.g. CLIP-seq/RIP-seq). Various sRNA library preparation protocols exist and during the training advantages and pitfalls of these methods will be discussed. Specific adaptations required for different types of sRNA will be highlighted. A practical course will teach the participants how to prepare sRNA NGS libraries themselves and how to perform quality control. Together, this training will enable participants to prepare sRNA libraries themselves autonomously, to validate the quality of their libraries and to perform successful NGS experiments by being cautious of technical pitfalls. More information: https://cnrsformation.cnrs.fr/preparer-des-banques-ngs-a-partir-des-petits-arn-pour-la-technologie-illumina

|
Scooped by
I2BC Paris-Saclay
September 20, 2023 5:31 AM
|
New arrivals on PIM facility
We are very pleased to welcome our colleague Stéphane Plancqueel (I2BC, crystallization platform) who joined the PIM team as a part time (20%) to help us with our growing activity. This arrival coincides with the launch of a new technology, a OMNISEC. This instrument is dedicated to the study of oligomerization states of proteins or complexes. Stéphane Plancqueel is an active member of Crystallisation facility and has recently joined the PIM facility to support the team with its growing activity. Stephane will complete his expertise on proteins by characterizing their oligomerization states and complexes, thanks to the different techniques present on PIM facility. He joined the PIM platform this summer as part time (20%) to strengthen the team and to work with Magali Aumont-Nicaise, Magali Noiray and Lia Maurin (M1 trainee).
In parallel, the OMNISEC was acquired thanks to supports from FRISBI and IBISA. It replaces the old SEC-MALS. Based on 2 different angles (RALS/LALS), the fully integrated OMNISEC system is a multi-detection, label-free, real-time analysis instrument. It is ideal for studying oligomerization states of proteins or to demonstrate their interactions. The OMNISEC system can be used for a wide range of analyses thanks to our Superdex chromatography columns from Cytiva. System provides up to 4 detections (UV, RI, RALS, LALS) in order to determine molecular sizes and cooling for temperature control down to 5°C in autosampler.

|
Scooped by
I2BC Paris-Saclay
September 20, 2023 5:08 AM
|
Workshop of Confocal microscopy
Annual workshop of confocal microscopy at Imagerie-Gif 9-13 October Subscriptions are open for the annual workshop of confocal microscopy at Imagerie-Gif on 9-13 October 2023. This workshop covers a large field of fluorescence imaging : from widefiled microscopy to super-resolution. It mixes theorical courses and large practical sessions. Beginners are welcome. For more information, link or please contact: romain.lebars@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
September 21, 2022 11:56 AM
|
Transmission Electron Microscopy Explore the architecture of a virus in all its forms
From October the 3rd to October the 10th Registration before September the 28th. More information here Contact: Claire Boulogne <claire.boulogne@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
July 25, 2022 4:53 AM
|
Training in Transmission electron microscopy for cell Biology
To acquire the theoretical basis of transmission electron microscopy (TEM) necessary for a functional exploration of the cell
Learn the different sample preparation techniques associated with TEM
Learn how to prepare your own sample Deadline registration September 27th. More information: here

|
Scooped by
I2BC Paris-Saclay
April 15, 2022 9:04 AM
|
Intensive week of training at Imagerie-Gif

|
Scooped by
I2BC Paris-Saclay
February 21, 2022 8:20 AM
|
Cryo-EM platform reveals first images of phages discovered by students
I2BC teams collaborate and create a teaching project to discover new phages with Bachelor students from Université Paris-Saclay. Financed by a grant from the University Paris-Saclay to develop teaching innovation, lecturers from I2BC’s Virology and Microbiology Departments have created a new lab course unit, called “Phage Discovery”, for second-year Bachelor students.
In September 2021, thirteen “phage hunters” signed up for this elective course and collected soil and water samples from the campus of Orsay and surroundings. Following purification, they isolated three distinct viruses that infect the bacterium Corynebacterium glutamicum. The students were welcomed by the cryo-electron microscopy platform of I2BC by Ana Arteni, Laura Pieri and Malika Ould Ali. Student phage hunters were dazzled by electron micrographs of their phage samples. Their day at the platform was the highlight of their first research experience.
The project will continue after the sequencing of the viral DNA by the I2BC platform: third-year Bachelor students will annotate the phage genomes. Stay tuned!
Contact: Ombeline Rossier, Christophe Regeard

|
Scooped by
I2BC Paris-Saclay
January 25, 2022 5:46 AM
|
First ICNS/I2BC Morning-Meeting February 8th at 9.30 a.m
The ICSN and the I2BC are the two major units of the CNRS campus of Gif sur Yvette. The “institut de Chimie des substances naturelles” (ICSN), with a staff of nearly 150 people, is the chemistry pole of the CNRS campus in Gif sur Yvette. This unit develops activities at the chemistry-biology interface, with natural substances as the object of study and main source of inspiration. Equipped with numerous platforms, including two NMR platforms, the ICSN will host an NMR apparatus from an I2BC team when it moves from the CEA-Saclay to the Gif-sur-Yvette campus. However, the links between our two institute are not limited to NMR; the purpose of this first morning is to introduce you to ICSN in the hope of encouraging new collaborations. This first Morning-Meeting, which will be the first of a series, will be oriented towards the platforms of the two units and duos of researchers to illustrate the fruitful collaborations already in place. Please come in large numbers to meet your chemical colleagues. Due to health conditions, the meeting will be held in hybrid mode, both in the auditorium of Building 21 on the Gif-sur-Yvette campus and on Zoom. See program on the website: here Contact for link: communication@i2bc.paris-saclay.fr
|

|
Scooped by
I2BC Paris-Saclay
April 3, 3:43 AM
|
Imagerie-Gif : New TIRF & Phase contrast modality

|
Scooped by
I2BC Paris-Saclay
February 10, 9:09 AM
|
Tutorial videos for ChimeraX

|
Scooped by
I2BC Paris-Saclay
December 17, 2024 11:56 AM
|
DciA, the Bacterial Replicative Helicase Loader, promotes LLPS in the presence of ssDNA.
This study shows that DciA, the bacterial replicative helicase loader, promotes condensates in the presence of DNA. This opens the way to the possibility of non-membrane compartments in bacteria. The goal could be to concentrate the players involved in replication and thus facilitate it. The loading of the bacterial replicative helicase DnaB is an essential step for genome replication and depends on the assistance of accessory proteins. Several of these proteins have been identified across the bacterial phyla. DciA is the most common loading protein in bacteria, yet the one whose mechanism is the least understood. We have previously shown that DciA from Vibrio cholerae is composed of a globular domain followed by an unfolded extension and demonstrated its strong affinity for DNA. Here, drawing on the skills of two I2BC facilities, Light microscopy (Imagerie-Gif) and PIM (Structural biology), we characterize the condensates formed by VcDciA upon interaction with a short single-stranded DNA substrate. We demonstrate the fluidity of these condensates using light microscopy and address their network organization through electron microscopy, thereby bridging events to conclude on a liquid-liquid phase separation behavior. Additionally, we observe the recruitment of DnaB in the droplets, concomitant with the release of DciA. We show that the well-known helicase loader DnaC from Escherichia coli is also competent to form these phase-separated condensates in the presence of ssDNA. Our phenomenological data are still preliminary as regards the existence of these condensates in vivo, but open the way for exploring the potential involvement of DciA in the formation of non-membrane compartments within the bacterium to facilitate the assembly of replication players on chromosomal DNA. More information : https://pubmed.ncbi.nlm.nih.gov/39603490/ Contact : Sophie CHERUEL sophie.quevillon-cheruel@i2bc.paris-saclay.fr and Stéphanie MARSIN stephanie.marsin@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
April 15, 2024 8:50 AM
|
A high-end Glacios2 cryo-electron microscope at I2BC
A Glacios2 200 kV has been delivered to I2BC, heralding, together with a Titan Krios 300 kV at synchrotron SOLEIL, a new era for cryo-electron microscopy in the Paris-Saclay area Cryo-electron microscopy (cryo-EM) has become an indispensable tool in structural biology, enabling researchers to unravel the complexities of biological macromolecules and understand their roles in various cellular processes and diseases. These last ten years the field has undergone a revolution, with technical advances that have made atomic resolution reconstructions of large macromolecular complexes possible (the so-called 'resolution revolution'), I2BC received the latest generation 200kV microscope, Glacios2, on March 27, 2024. Joint operation is planned with the 300kV Titan Krios also just delivered at synchrotron SOLEIL. This will allow researchers to study structures of living machines at work in fine detail, both for purified and/or reconstituted complexes and in cells. These two cryo-electron microscopes are gathering scientists from all over the Paris-Saclay area, sparking further interest in future developments at the interface beween structural and cellular biology. contact: Stéphane Bressanelli (plt-cryoem@i2bc.paris-saclay.fr)

|
Scooped by
I2BC Paris-Saclay
December 27, 2023 9:21 AM
|
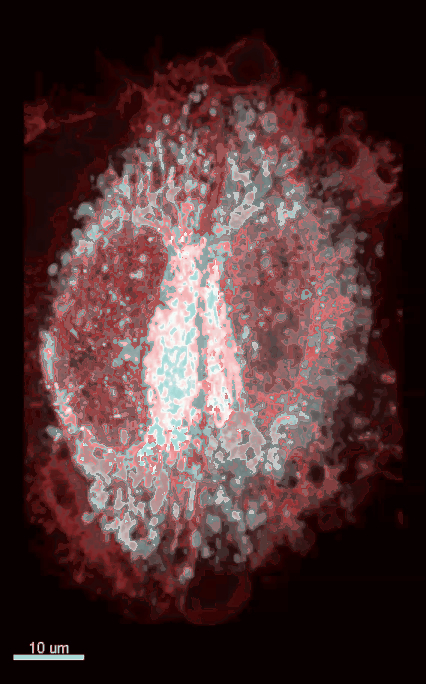
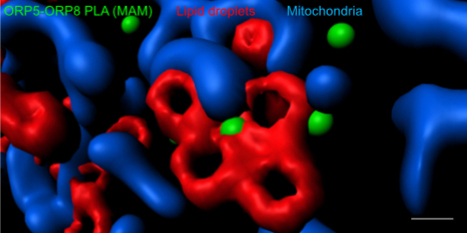
Visualization and Quantification of Endogenous Intra-Organelle Protein Interactions at ER-Mitochondria Contact Sites by Proximity Ligation Assays

|
Scooped by
I2BC Paris-Saclay
October 23, 2023 7:12 AM
|
New arrival at the BIOI2 bioinformatics plateform
Fadwa EL KHADDAR is working with us part time at the BIOI2 plateform since the beginning of October 2023 and has brought with her leading-edge experience in omics data analysis that will greatly enrich the range of services offered by the plateform. We welcome Fadwa EL KHADDAR, an engineer in bioinformatics who will be helping out part time (20%) at the BIOI2 plateform, as well as integrating two research teams of the I2BC (BIM and SSFA of the Genomes department) to which she will be lending her expertise in Omics data analysis. Fadwa first discovered bioinformatics during her Master's degree in Medical biotechnology of Rabat, Morocco, where she worked on phylogeny, synteny et pangenome analysis of L. monocytogenes bacteria as an intern in the Biotechnology & bioresource valorisation laboratory (Meknes, Morocco). She more recently followed a Master's degree in Bioinformatics in Montpellier, where she integrated the AGAP Institute (CIRAD, Montpellier) as a trainee and developed an identification pipeline for gene resistance families (NBS-LRR) in common grape vine, V. vinifera. She then completed her Master thesis in the IRD (Montpellier) working on raw long-read nanopore sequencing data to detect whole genome cytosine methylation in model and orphan plant species. With her know-how, Fadwa will be nicely completing the range of expertise offered by the BIOI2 plateform. BIOI2 provides access to bioinformatics resources and activities developed and of use at the I2BC, organises focused training session on bioinformatics tools, and is an official contributing plateform of the nationwide French Institute of Bioinformatics (IFB).

|
Scooped by
I2BC Paris-Saclay
September 20, 2023 5:11 AM
|
Workshop of Electron Microscopy
Annual workshop of electron microscopy at Imagerie-Gif 17-20 October 2023 Subscriptions are open for the annual workshop of electron microscopy at Imagerie-Gif on 17-20 October 2023. This workshop covers all sample preparation techniques for cell biology in electron microscopy. It mixes theorical courses and large practical sessions. Beginners are welcome.
For more information link or please contact: claire.boulogne@i2bc.paris-saclay.fr

|
Scooped by
I2BC Paris-Saclay
March 17, 2023 11:49 AM
|
First heliX installed in France
Studying molecular interactions of nucleic acid binders, affinity vs. avidity, and ternary complexes with the heliX+ biosensor. The I2BC PIM platform proposed the swithSENSE technology since 2017 that uses nanolevers bound on a gold surface to measure protein interactions in an approach highly complementary to other surface methods like BioLayer Interferometry and Surface Plasmon Resonance also available on PIM platform (https://www.i2bc.paris-saclay.fr/structural-biology/protein-interaction/). In January 2023, the new generation switchSENSE instrument, heliX+ was installed in place of the first generation DRX2. This is the first instrument implemented in France. It has unique features in particular for DNA/RNA Binding Proteins. It can resolve the fastest kinetics with confidence using advanced microfluidics and 10 ms data collection (Nemoz, 2018, NSMB, Velours, 2021, Eur Bioph Journal). Molecular interactions can be detected with femto-molar sensitivity. It allows screening and ranking of small molecule induced conformational changes (96 and 384 well plates). It allows to follow enzymatic activities including polymerases, ligases and nucleases.

|
Scooped by
I2BC Paris-Saclay
September 21, 2022 10:18 AM
|
Confocal Microscopy Workshop (October 10-14)
Acquire and reinforce the theoretical basis of light microscopy
Deepen and master the practical use of a wide field and confocal microscope
Understand the complementarities of techniques from conventional microscopy to super-resolution. Registration deadline September 27th here Contact: Romain Le Bars <romain.lebars@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
June 15, 2022 11:32 AM
|
Interdisciplinary access call to Structural biology, Biological imaging and Proteomics IBISA facilities
The 3 national infrastructures ProFi, FBI and FRISBI along with the GIS IBiSA proposed a call for a funded access to IBiSA-labelled facilities. This 1st call was a success with 7 projects selected among which one using the Imagerie-Gif facility (@I2BC). The three national infrastructures ProFi, FranceBioImaging and FRISBI along with the GIS IBiSA proposed a call for a funded access to IBiSA-labelled facilities. The aim was to promote IBiSA facilities networking through transdisciplinary research projects. This 1st call was a success with 26 proposals submitted. Seven projects were selected among which one using using the Imagerie-Gif facility.
One project coordinated by Mireille Bétermier in collaboration with JB Charbonnier was selected. It includes access to the Cytometry platform of Imagerie-Gif at I2BC (France BioImaging), the Proteopole Biophysics platform of Institut Pasteur and the ProteoSeine Mass spectrometry platform of Institut Jacques Monod. The project aims at using approaches from these three IBiSA platforms to integrate complementary data on programmed genome rearrangement and double-strand break repair in Paramecium Contact: Julie Ménétrey <julie.menetrey@i2bc.paris-saclay.fr>

|
Scooped by
I2BC Paris-Saclay
March 1, 2022 4:58 AM
|
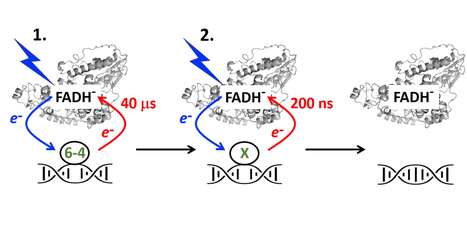
Kinetics of electron returns in the two-photon DNA repair by (6-4) photolyase
DNA repairing enzymes photolyases are ubiquitous natural catalysts using sunlight to revert cancerogenic chemical changes in DNA caused by UV light. The 3P Team Photobiology, Photosynthesis, Photocatalysis from the Institute of Integrative Biology of the Cell (I2BC) explores the molecular mechanism by which these essential photoenzymes have been repairing DNA since the beginnings of life evolution on Earth. In their latest work published in the journal ACS Catalysis, the researchers have shown that the repair of the UV-induced lesions called ‘(6-4) photoproducts’ occurs in two successive photoreactions (requiring absorption of two separate photons), each beginning with electron transfer from the excited flavin to the lesion. Transient absorption spectroscopy (https://www.pluginlabs-universiteparissaclay.fr/fr/entity/a5ad9e5d-53a4-4194-8653-3d8feebfafc8/i2bc-plateforme-de-spectroscopies-electroniques) was used to follow the return of electrons to the flavin after chemical transformations of the lesion. The team was able to dissect the electron return kinetics finalizing the first and the second photoreactions (in ~40 μs and ~200 ns, respectively), corroborating and detailing the two-photon reaction model. More information: https://doi.org/10.1021/acscatal.2c00492 Authors: Klaus Brettel, Pavel Müller (I2BC) and Junpei Yamamoto (Osaka University). The project was funded by the French National Research Agency (ANR). Contact: Pavel Müller <pavel.muller@i2bc.paris-saclay.fr.>

|
Scooped by
I2BC Paris-Saclay
February 21, 2022 7:47 AM
|
3-days Training in Flow Cytometry
Imagerie-Gif and CNRS Formation Entreprises are organizing a training to blow your mind with flow cytometry. We offer you to acquire the theoretical bases in flow cytometry (analysis and sorting, experimental strategies), to be informed of applications and developments in very varied fields of application, and to initiate or deepen the use of analyzers and sorters.
The training will be split in theoretical mornings and practical afternoons. Practical afternoons will take place on GIF and ORSAY campus.
The training is opened to Student, researcher and Engineer. https://cnrsformation.cnrs.fr/atelier-cytometrie?mc=Cytom%C3%A9trie Contact person: BOURGE Mickael
|




 Your new post is loading...
Your new post is loading...